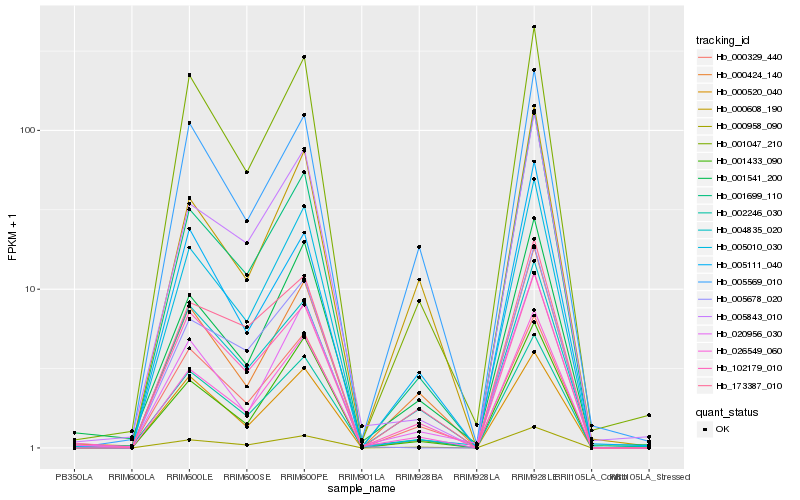
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000958_090 |
0.0 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 2 |
Hb_026549_060 |
0.0643206525 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 3 |
Hb_000424_140 |
0.0666672942 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_001047_210 |
0.0715568303 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 5 |
Hb_004835_020 |
0.0718431559 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 6 |
Hb_173387_010 |
0.0763292993 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 7 |
Hb_005569_010 |
0.0791318111 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 8 |
Hb_001699_110 |
0.0793635786 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 9 |
Hb_102179_010 |
0.0825109128 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_005010_030 |
0.0835998881 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 11 |
Hb_000608_190 |
0.0906341544 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 12 |
Hb_000520_040 |
0.0917563843 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 13 |
Hb_000329_440 |
0.0918513764 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 14 |
Hb_005843_010 |
0.0933137999 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 15 |
Hb_002246_030 |
0.0935207662 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_001433_090 |
0.0946929575 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 17 |
Hb_005111_040 |
0.097578562 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_020956_030 |
0.0978450312 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_001541_200 |
0.0997755636 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_005678_020 |
0.1002021388 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |