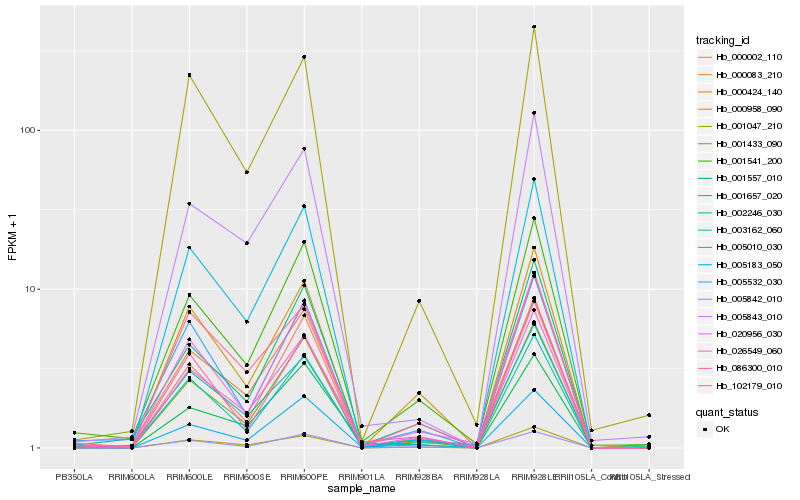
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026549_060 |
0.0 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 2 |
Hb_001433_090 |
0.0541052426 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 3 |
Hb_001541_200 |
0.061060609 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_000958_090 |
0.0643206525 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 5 |
Hb_020956_030 |
0.0692829924 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 6 |
Hb_005183_050 |
0.0769686197 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 7 |
Hb_005010_030 |
0.0793248577 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 1 [Jatropha curcas] |
| 8 |
Hb_001657_020 |
0.0800488687 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 9 |
Hb_002246_030 |
0.0839172176 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 10 |
Hb_003162_060 |
0.0843146415 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 11 |
Hb_001047_210 |
0.0861468005 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 12 |
Hb_000424_140 |
0.0870097556 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_005843_010 |
0.0881070176 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 14 |
Hb_086300_010 |
0.0884751767 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 15 |
Hb_000002_110 |
0.0900799165 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 16 |
Hb_001557_010 |
0.090507236 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 17 |
Hb_005532_030 |
0.0935795522 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_005842_010 |
0.0937076511 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_000083_210 |
0.0955867912 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 20 |
Hb_102179_010 |
0.0957431844 |
- |
- |
protein binding protein, putative [Ricinus communis] |