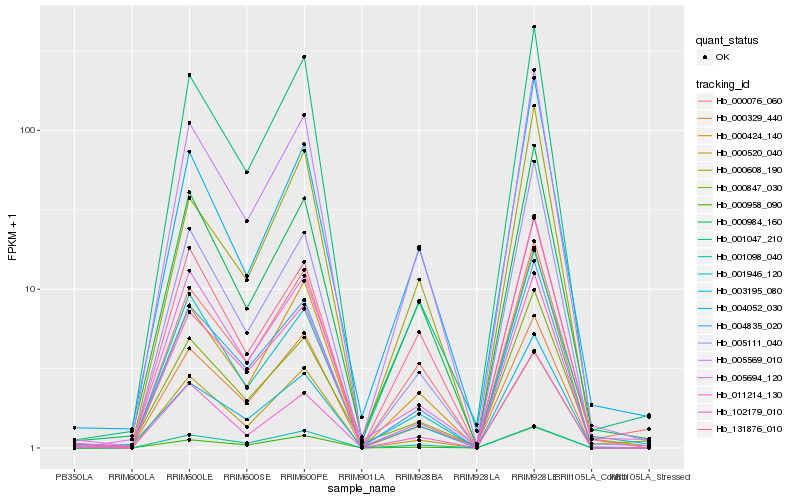
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005569_010 |
0.0 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 2 |
Hb_004835_020 |
0.0469109468 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 3 |
Hb_000984_160 |
0.0533349119 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000424_140 |
0.0558732895 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_003195_080 |
0.0716037348 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 6 |
Hb_000329_440 |
0.0789899554 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 7 |
Hb_000958_090 |
0.0791318111 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 8 |
Hb_000076_060 |
0.0825442943 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 9 |
Hb_102179_010 |
0.0831624635 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_005694_120 |
0.0847673772 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 11 |
Hb_000520_040 |
0.0850712885 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 12 |
Hb_000847_030 |
0.0865867326 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 13 |
Hb_131876_010 |
0.0879084509 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_005111_040 |
0.0879353345 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_001098_040 |
0.0892429954 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 16 |
Hb_001946_120 |
0.0893951896 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_004052_030 |
0.0899644144 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 18 |
Hb_011214_130 |
0.0918652746 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 19 |
Hb_001047_210 |
0.0925100148 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 20 |
Hb_000608_190 |
0.0967056926 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |