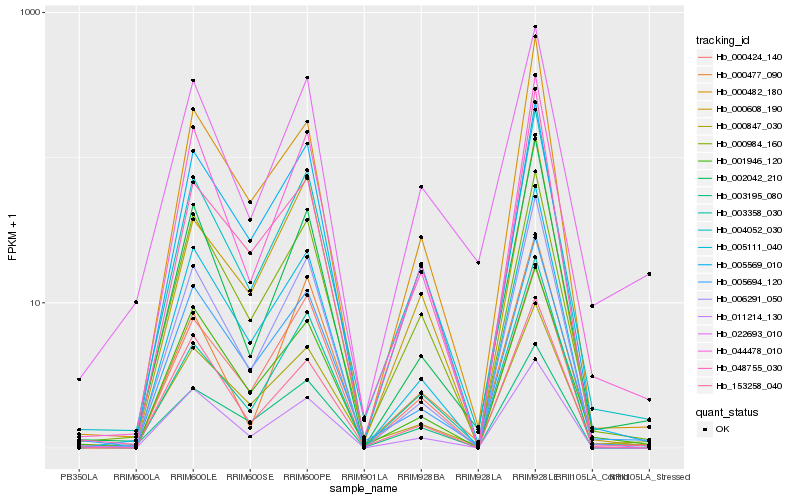
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 2 |
Hb_000984_160 |
0.0704709089 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_044478_010 |
0.0761426298 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 4 |
Hb_003358_030 |
0.0829168498 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 5 |
Hb_005111_040 |
0.0861379645 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_005569_010 |
0.0899644144 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 7 |
Hb_000482_180 |
0.0909251148 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 8 |
Hb_005694_120 |
0.0920056834 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 9 |
Hb_000608_190 |
0.0923961463 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 10 |
Hb_000847_030 |
0.0929094936 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 11 |
Hb_002042_210 |
0.0947034289 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003195_080 |
0.0975060002 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 13 |
Hb_006291_050 |
0.0992848465 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_001946_120 |
0.1001039973 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 15 |
Hb_022693_010 |
0.1006870722 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 16 |
Hb_153258_040 |
0.1013941376 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 17 |
Hb_011214_130 |
0.1027992024 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 18 |
Hb_000424_140 |
0.1029474347 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_048755_030 |
0.1075043568 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000477_090 |
0.1101543932 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |