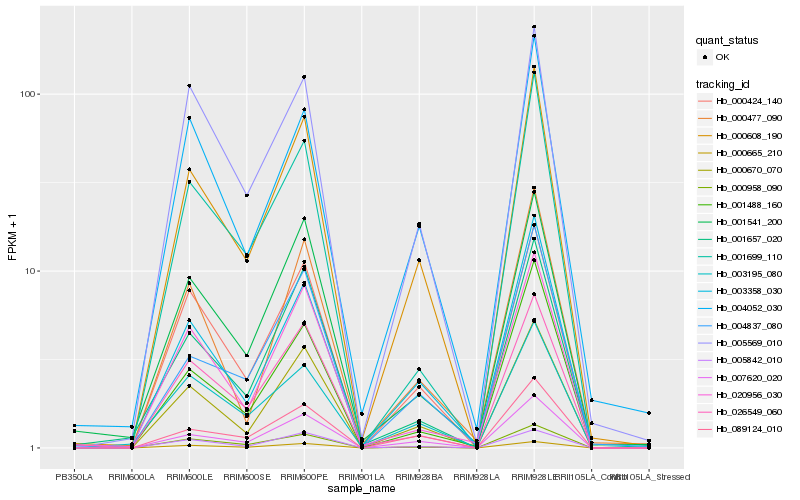
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_190 |
0.0 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 2 |
Hb_007620_020 |
0.0557580186 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000670_070 |
0.0581516484 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 4 |
Hb_000424_140 |
0.0685587873 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_003358_030 |
0.0712221626 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 6 |
Hb_089124_010 |
0.072353154 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 7 |
Hb_000958_090 |
0.0906341544 |
- |
- |
PREDICTED: cation/H(+) antiporter 28 [Jatropha curcas] |
| 8 |
Hb_004052_030 |
0.0923961463 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 9 |
Hb_004837_080 |
0.0934850251 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 10 |
Hb_005569_010 |
0.0967056926 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 11 |
Hb_003195_080 |
0.0975737196 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 12 |
Hb_001541_200 |
0.098124721 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 13 |
Hb_026549_060 |
0.0988131261 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_001657_020 |
0.0994663918 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 15 |
Hb_001699_110 |
0.1042844831 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 16 |
Hb_001488_160 |
0.1043594808 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000477_090 |
0.1055027832 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 18 |
Hb_020956_030 |
0.1058805291 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_005842_010 |
0.1080264365 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 20 |
Hb_000665_210 |
0.1085768883 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |