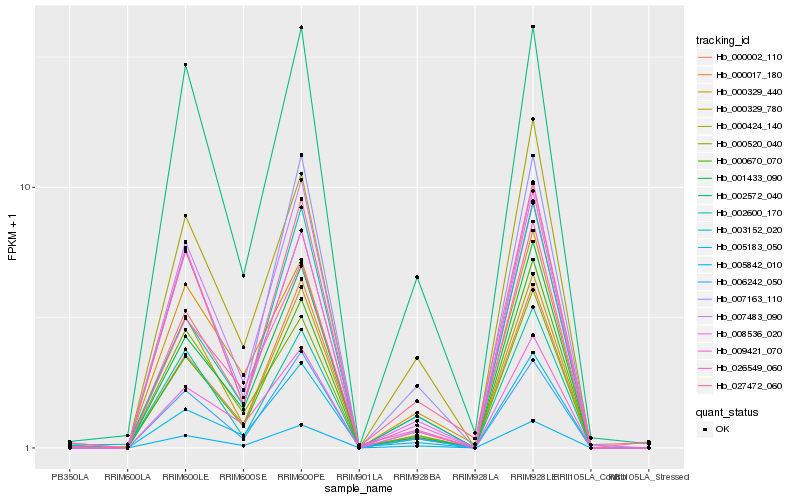
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005842_010 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_006242_050 |
0.0650320325 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 3 |
Hb_000424_140 |
0.0658843536 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_007163_110 |
0.069195005 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 5 |
Hb_000670_070 |
0.0727510381 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 6 |
Hb_009421_070 |
0.0728259575 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 7 |
Hb_027472_060 |
0.0756138742 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 8 |
Hb_005183_050 |
0.0777089005 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 9 |
Hb_000329_780 |
0.0816405162 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 10 |
Hb_001433_090 |
0.0820166691 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 11 |
Hb_000017_180 |
0.0849519106 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 12 |
Hb_007483_090 |
0.0864185261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008536_020 |
0.0874805541 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 14 |
Hb_000002_110 |
0.0888051082 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 15 |
Hb_000520_040 |
0.0888794024 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 16 |
Hb_002572_040 |
0.0896638863 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 17 |
Hb_002600_170 |
0.0903398429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000329_440 |
0.0930030044 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 19 |
Hb_003152_020 |
0.093451978 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 20 |
Hb_026549_060 |
0.0937076511 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |