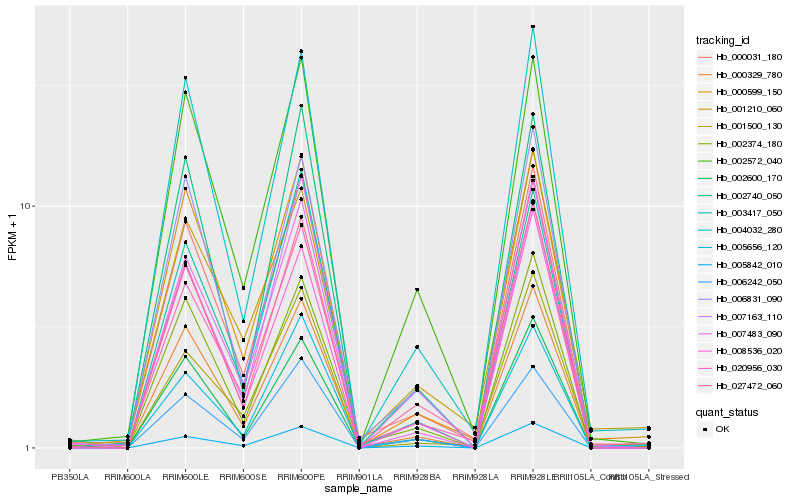
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027472_060 |
0.0 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 2 |
Hb_004032_280 |
0.0625778048 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 3 |
Hb_000031_180 |
0.0647856071 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 4 |
Hb_005842_010 |
0.0756138742 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 5 |
Hb_000329_780 |
0.0757575383 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 6 |
Hb_002572_040 |
0.077535772 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 7 |
Hb_000599_150 |
0.0786869332 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 8 |
Hb_006831_090 |
0.080155053 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002600_170 |
0.0811985939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006242_050 |
0.086261086 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 11 |
Hb_007163_110 |
0.0867583494 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 12 |
Hb_008536_020 |
0.0891822792 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 13 |
Hb_002374_180 |
0.0900230175 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 14 |
Hb_005656_120 |
0.0936623019 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 15 |
Hb_001210_060 |
0.0946631497 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 16 |
Hb_002740_050 |
0.0954552897 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 17 |
Hb_020956_030 |
0.0972479638 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 18 |
Hb_003417_050 |
0.0974210392 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001500_130 |
0.0975033302 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 20 |
Hb_007483_090 |
0.0978417947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |