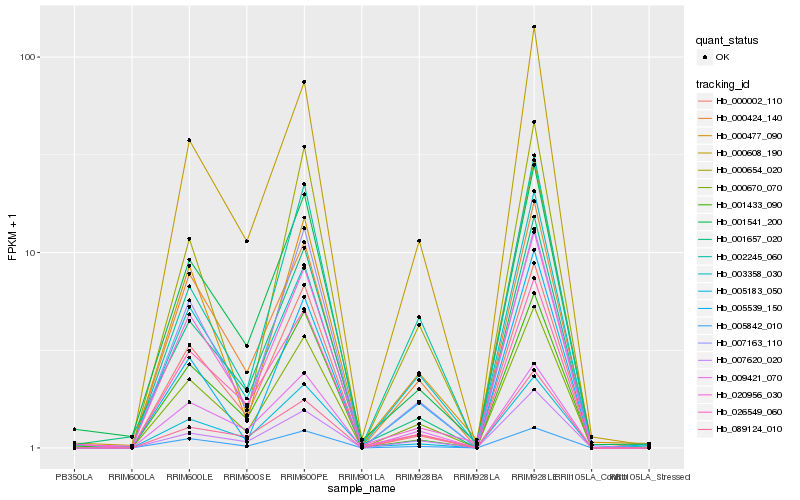
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000670_070 |
0.0 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 2 |
Hb_000608_190 |
0.0581516484 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 3 |
Hb_000424_140 |
0.0642395669 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_007620_020 |
0.0702531709 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005842_010 |
0.0727510381 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_000654_020 |
0.0847521892 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 7 |
Hb_002245_060 |
0.0849363017 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000477_090 |
0.0940022889 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 9 |
Hb_001657_020 |
0.094451094 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 10 |
Hb_000002_110 |
0.0958401819 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 11 |
Hb_020956_030 |
0.0958628377 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 12 |
Hb_005539_150 |
0.0965370581 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 13 |
Hb_005183_050 |
0.096578645 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 14 |
Hb_003358_030 |
0.0981766663 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 15 |
Hb_009421_070 |
0.0982884692 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 16 |
Hb_001541_200 |
0.0996630644 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 17 |
Hb_007163_110 |
0.1001016551 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_089124_010 |
0.1007796412 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 19 |
Hb_001433_090 |
0.1011673949 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 20 |
Hb_026549_060 |
0.1043336909 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |