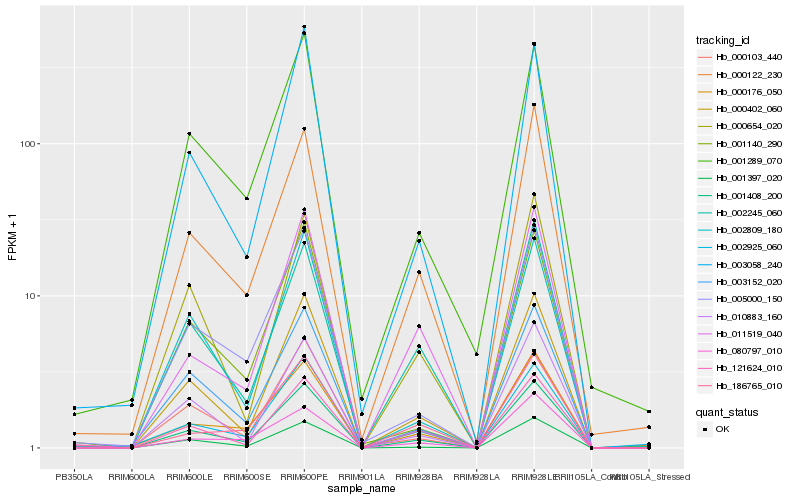
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_200 |
0.0 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 2 |
Hb_000402_060 |
0.0513105982 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002925_060 |
0.0735638294 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 4 |
Hb_000122_230 |
0.0759044935 |
- |
- |
laccase, putative [Ricinus communis] |
| 5 |
Hb_003152_020 |
0.0806086159 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 6 |
Hb_001397_020 |
0.0837237346 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 7 |
Hb_003058_240 |
0.0850653786 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 8 |
Hb_010883_160 |
0.0862807647 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002245_060 |
0.087370141 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005000_150 |
0.0889354284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001289_070 |
0.0921351687 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 12 |
Hb_000654_020 |
0.096514488 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 13 |
Hb_011519_040 |
0.0980823705 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 14 |
Hb_002809_180 |
0.0996166755 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 15 |
Hb_000103_440 |
0.0997030865 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 16 |
Hb_080797_010 |
0.1022475471 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570-like [Citrus sinensis] |
| 17 |
Hb_001140_290 |
0.103834789 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 18 |
Hb_000176_050 |
0.1044337942 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_121624_010 |
0.1072931814 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 20 |
Hb_186765_010 |
0.1080246039 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |