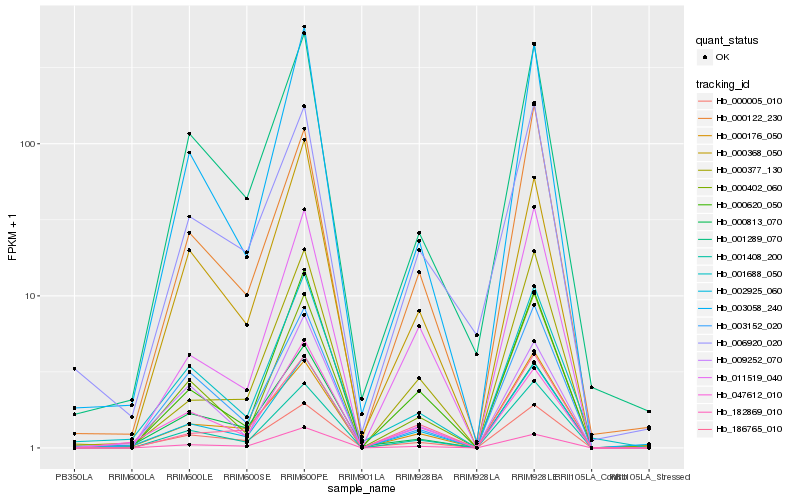
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002925_060 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 2 |
Hb_001408_200 |
0.0735638294 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 3 |
Hb_000620_050 |
0.077458685 |
- |
- |
PREDICTED: uncharacterized protein LOC105649924 [Jatropha curcas] |
| 4 |
Hb_182869_010 |
0.0836901749 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_003058_240 |
0.089438421 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 6 |
Hb_047612_010 |
0.0948099629 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_000402_060 |
0.0950937655 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001289_070 |
0.0978965512 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_001688_050 |
0.0982052907 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 10 |
Hb_011519_040 |
0.0982517181 |
- |
- |
hypothetical protein POPTR_0015s04770g [Populus trichocarpa] |
| 11 |
Hb_186765_010 |
0.1000819076 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 12 |
Hb_000377_130 |
0.1015618887 |
- |
- |
hypothetical protein POPTR_0016s00790g [Populus trichocarpa] |
| 13 |
Hb_000813_070 |
0.1044325294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000368_050 |
0.1058314706 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 15 |
Hb_000176_050 |
0.1079791457 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_003152_020 |
0.1102342129 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 17 |
Hb_006920_020 |
0.1105687851 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 18 |
Hb_000005_010 |
0.1153115742 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 19 |
Hb_000122_230 |
0.1159300227 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_009252_070 |
0.1161735984 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |