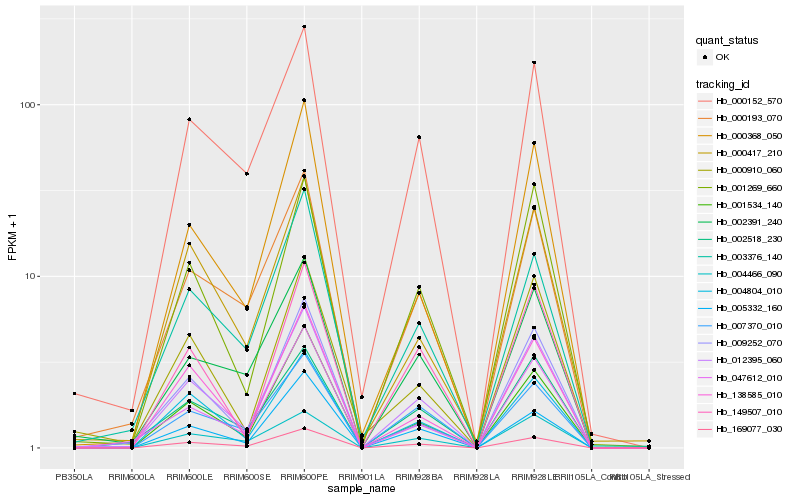
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012395_060 |
0.0 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 2 |
Hb_169077_030 |
0.0696495487 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 3 |
Hb_004804_010 |
0.0801021596 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 4 |
Hb_001269_660 |
0.0834088906 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 5 |
Hb_001534_140 |
0.0863319594 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 6 |
Hb_138585_010 |
0.0912781157 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 7 |
Hb_000152_570 |
0.0928818958 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 8 |
Hb_000193_070 |
0.093006935 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 9 |
Hb_002518_230 |
0.0937130099 |
- |
- |
- |
| 10 |
Hb_149507_010 |
0.0970523837 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Malus domestica] |
| 11 |
Hb_047612_010 |
0.0980087285 |
- |
- |
glutamine amidotransferase class-I domain-containing family protein [Populus trichocarpa] |
| 12 |
Hb_000368_050 |
0.1017597207 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_009252_070 |
0.1029565777 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 14 |
Hb_000417_210 |
0.1047683124 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002391_240 |
0.1057271767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007370_010 |
0.1058088535 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 17 |
Hb_004466_090 |
0.1058675575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003376_140 |
0.1060229197 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 19 |
Hb_005332_160 |
0.1074327849 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 20 |
Hb_000910_060 |
0.1080629249 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |