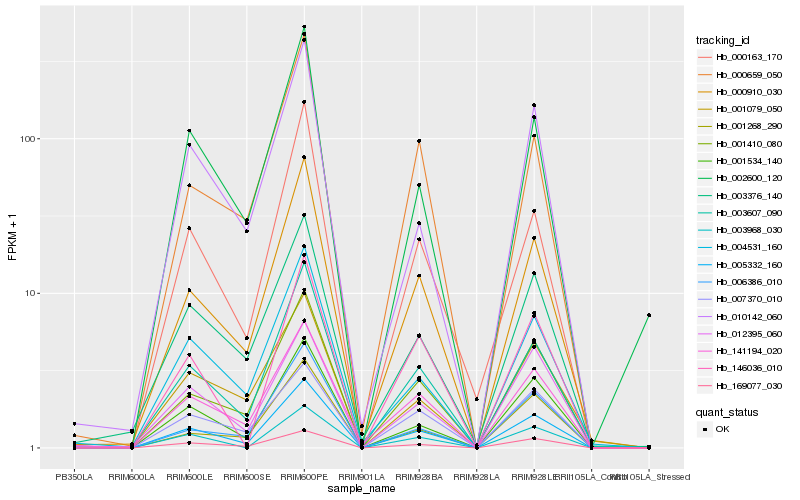
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_160 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 2 |
Hb_003607_090 |
0.061219805 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 3 |
Hb_000910_030 |
0.0676821584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001534_140 |
0.0730042309 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 5 |
Hb_001410_080 |
0.0782090831 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_169077_030 |
0.0811134213 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 7 |
Hb_141194_020 |
0.0875002269 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 8 |
Hb_004531_160 |
0.0943333453 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003376_140 |
0.1004321452 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 10 |
Hb_002600_120 |
0.1040639002 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 11 |
Hb_010142_060 |
0.1056970556 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 12 |
Hb_012395_060 |
0.1074327849 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 13 |
Hb_000163_170 |
0.1083192388 |
- |
- |
- |
| 14 |
Hb_007370_010 |
0.1122215207 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 15 |
Hb_003968_030 |
0.1129565802 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_006386_010 |
0.1136370015 |
- |
- |
PREDICTED: polygalacturonase QRT3 [Jatropha curcas] |
| 17 |
Hb_001268_290 |
0.1159893075 |
- |
- |
PREDICTED: glutaredoxin-C1-like [Jatropha curcas] |
| 18 |
Hb_001079_050 |
0.1174928347 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 19 |
Hb_000659_050 |
0.1185943506 |
- |
- |
PREDICTED: basic blue protein-like [Jatropha curcas] |
| 20 |
Hb_146036_010 |
0.1202291022 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |