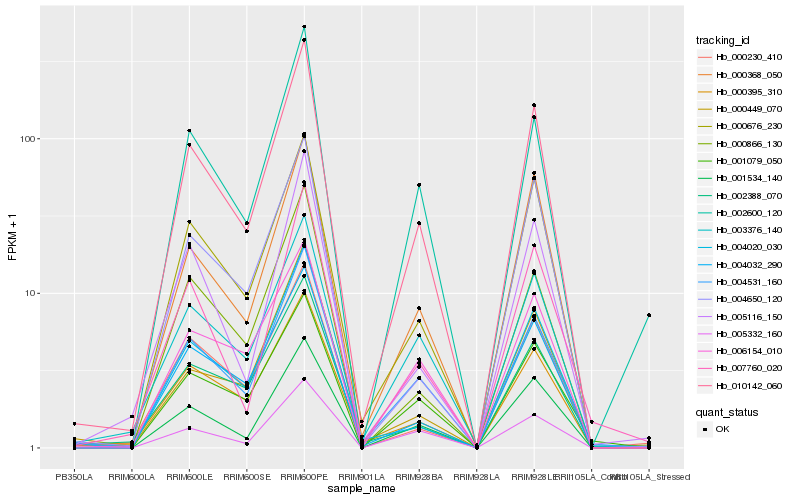
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010142_060 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_001534_140 |
0.0626066492 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 3 |
Hb_004531_160 |
0.0653630535 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000676_230 |
0.070133797 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 5 |
Hb_000368_050 |
0.0814404685 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 6 |
Hb_004032_290 |
0.0847963376 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000395_310 |
0.0862673154 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_004020_030 |
0.0905161745 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 9 |
Hb_000866_130 |
0.0946640155 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 10 |
Hb_003376_140 |
0.0958229916 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 11 |
Hb_005116_150 |
0.096572767 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002600_120 |
0.0996264074 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 13 |
Hb_007760_020 |
0.1008746604 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 14 |
Hb_001079_050 |
0.1028854677 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_002388_070 |
0.1037471672 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_006154_010 |
0.105296206 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000449_070 |
0.1055319353 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_005332_160 |
0.1056970556 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 19 |
Hb_004650_120 |
0.1057385895 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 20 |
Hb_000230_410 |
0.1058119649 |
- |
- |
PREDICTED: uncharacterized protein LOC105631891 [Jatropha curcas] |