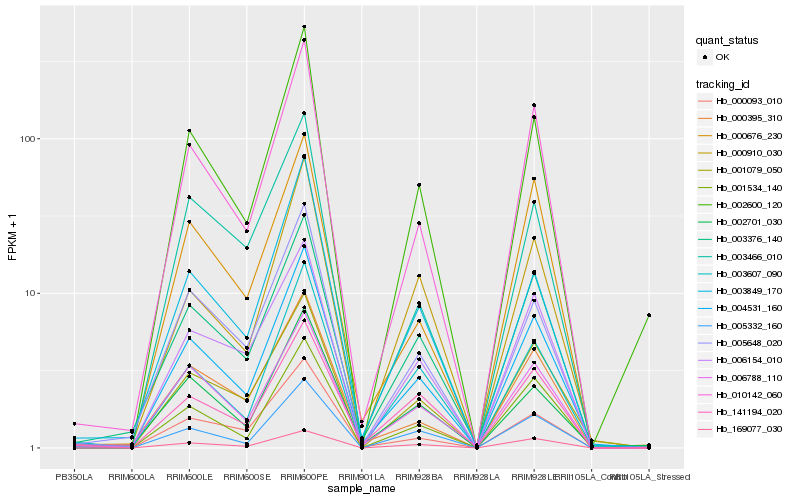
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_160 |
0.0 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_010142_060 |
0.0653630535 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_003376_140 |
0.0724878562 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 4 |
Hb_001079_050 |
0.0892376971 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 5 |
Hb_003607_090 |
0.0902198592 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 6 |
Hb_002600_120 |
0.0906413175 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 7 |
Hb_005648_020 |
0.0918035545 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 8 |
Hb_001534_140 |
0.0927161968 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 9 |
Hb_000395_310 |
0.0928781277 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_005332_160 |
0.0943333453 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 2-like [Jatropha curcas] |
| 11 |
Hb_000093_010 |
0.0969406126 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 12 |
Hb_002701_030 |
0.098129279 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 13 |
Hb_006788_110 |
0.099053409 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 14 |
Hb_006154_010 |
0.0997431751 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000910_030 |
0.1019936254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003849_170 |
0.1034826496 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 17 |
Hb_141194_020 |
0.1053482065 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 18 |
Hb_169077_030 |
0.1087389843 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 19 |
Hb_000676_230 |
0.1092110032 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 20 |
Hb_003466_010 |
0.1113129336 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |