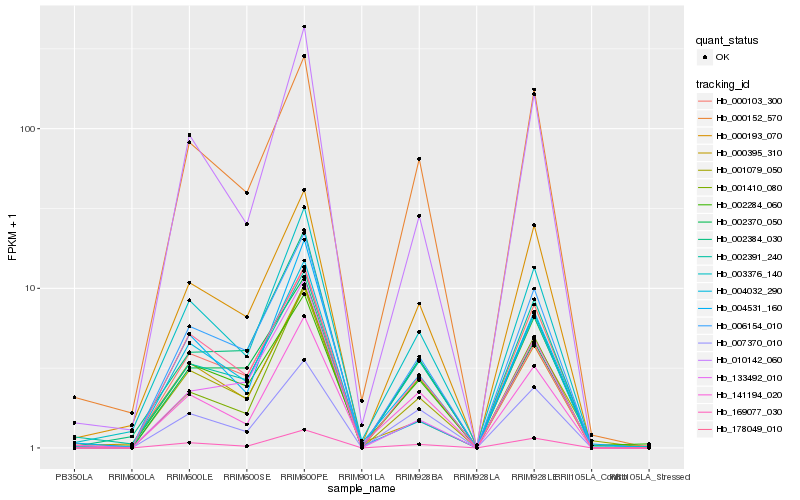
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006154_010 |
0.0 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001079_050 |
0.0661436777 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 3 |
Hb_000103_300 |
0.0770930414 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 4 |
Hb_000193_070 |
0.0775798387 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 5 |
Hb_003376_140 |
0.077847644 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 6 |
Hb_169077_030 |
0.0800638821 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 7 |
Hb_000152_570 |
0.0918358873 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 8 |
Hb_002384_030 |
0.0973699033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004531_160 |
0.0997431751 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_178049_010 |
0.1025379223 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002370_050 |
0.1026652294 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 12 |
Hb_002391_240 |
0.1042215743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010142_060 |
0.105296206 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 14 |
Hb_002284_060 |
0.1083356602 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 15 |
Hb_000395_310 |
0.1092250571 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_001410_080 |
0.1094120886 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_133492_010 |
0.11094937 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_141194_020 |
0.1137978546 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 19 |
Hb_004032_290 |
0.1139321342 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_007370_010 |
0.1147099266 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |