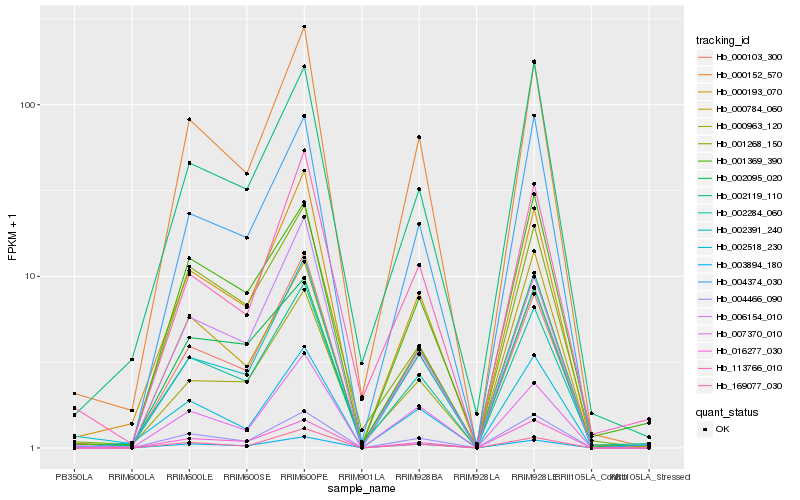
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_060 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_000103_300 |
0.0578768875 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 3 |
Hb_000152_570 |
0.0591001192 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 4 |
Hb_000193_070 |
0.0696458101 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 5 |
Hb_004466_090 |
0.0875130305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002391_240 |
0.0907609747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003894_180 |
0.0924750155 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 8 |
Hb_004374_030 |
0.0979915893 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 9 |
Hb_001268_150 |
0.0981583778 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_001369_390 |
0.0996091452 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 11 |
Hb_113766_010 |
0.1028580628 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 12 |
Hb_002095_020 |
0.1048876442 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_016277_030 |
0.1051540597 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 14 |
Hb_169077_030 |
0.1060397836 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 15 |
Hb_002119_110 |
0.1076817827 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 28 [Jatropha curcas] |
| 16 |
Hb_006154_010 |
0.1083356602 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_007370_010 |
0.1099088554 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 18 |
Hb_000963_120 |
0.1100203589 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002518_230 |
0.1102886003 |
- |
- |
- |
| 20 |
Hb_000784_060 |
0.1113637216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |