| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
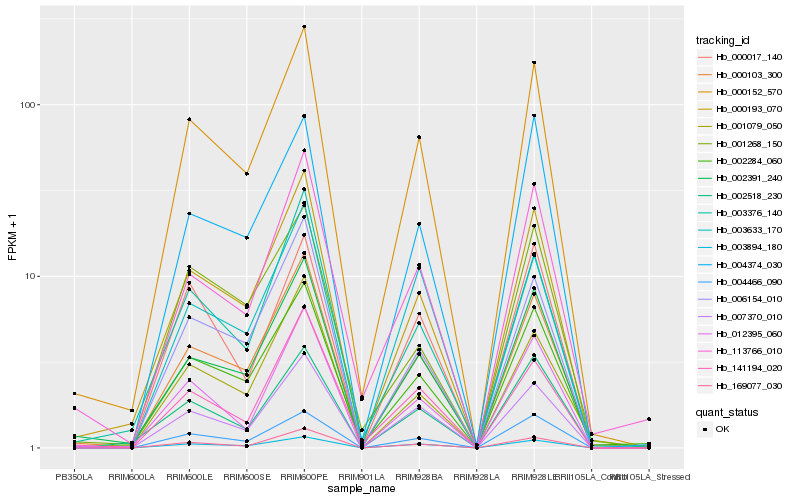
Hb_000152_570 |
0.0 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 2 |
Hb_000103_300 |
0.0474551771 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 3 |
Hb_000193_070 |
0.0482430818 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 4 |
Hb_002284_060 |
0.0591001192 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_002391_240 |
0.06755893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003894_180 |
0.0729014038 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 7 |
Hb_007370_010 |
0.0731154502 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 8 |
Hb_169077_030 |
0.0732493177 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 9 |
Hb_004466_090 |
0.0831274642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006154_010 |
0.0918358873 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_012395_060 |
0.0928818958 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 12 |
Hb_003376_140 |
0.0944705403 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 13 |
Hb_002518_230 |
0.0997705352 |
- |
- |
- |
| 14 |
Hb_141194_020 |
0.1009503451 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 15 |
Hb_113766_010 |
0.1016350218 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 16 |
Hb_004374_030 |
0.1076708034 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 17 |
Hb_001079_050 |
0.1100698796 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 18 |
Hb_000017_140 |
0.1131358435 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 19 |
Hb_001268_150 |
0.1150826414 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_003633_170 |
0.117657658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |