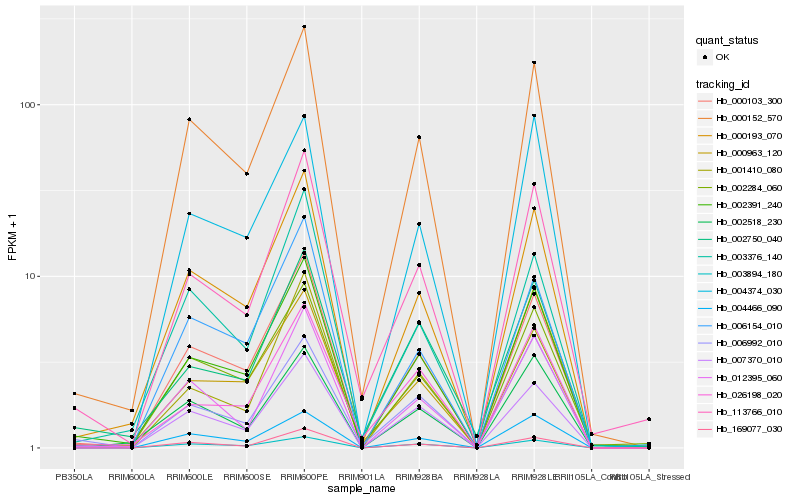
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000193_070 |
0.0557663127 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 3 |
Hb_000103_300 |
0.0634959116 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 4 |
Hb_000152_570 |
0.06755893 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 5 |
Hb_002284_060 |
0.0907609747 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 6 |
Hb_113766_010 |
0.0955588463 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 7 |
Hb_007370_010 |
0.0981707608 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 8 |
Hb_169077_030 |
0.102374804 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 9 |
Hb_004466_090 |
0.103607322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006154_010 |
0.1042215743 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_012395_060 |
0.1057271767 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 12 |
Hb_003894_180 |
0.1066824931 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 13 |
Hb_026198_020 |
0.1067561429 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 14 |
Hb_001410_080 |
0.1077132972 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 15 |
Hb_004374_030 |
0.108575848 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_002518_230 |
0.1088635837 |
- |
- |
- |
| 17 |
Hb_002750_040 |
0.1128509479 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 18 |
Hb_006992_010 |
0.11488516 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_003376_140 |
0.116904194 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 20 |
Hb_000963_120 |
0.1171024885 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |