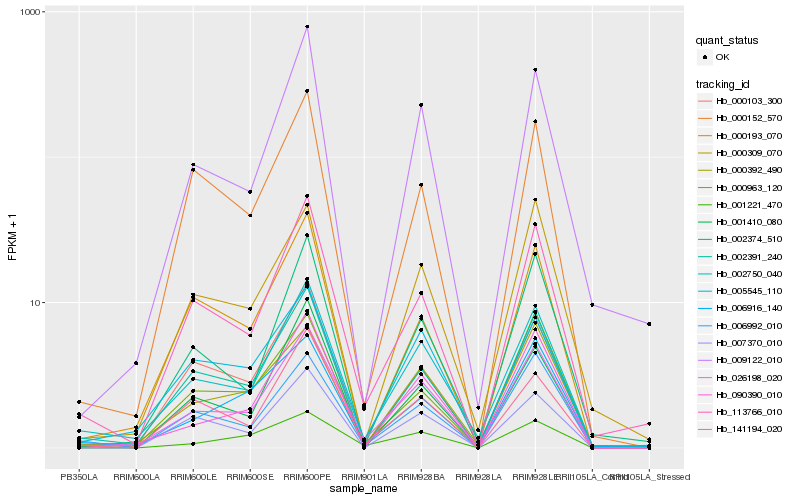
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026198_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 2 |
Hb_002750_040 |
0.082575296 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 3 |
Hb_113766_010 |
0.0979717637 |
- |
- |
plasma membrane H+ ATPase, partial [Nicotiana plumbaginifolia] |
| 4 |
Hb_002391_240 |
0.1067561429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_090390_010 |
0.1078302371 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 6 |
Hb_000392_490 |
0.1146846711 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006992_010 |
0.1150505506 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_009122_010 |
0.1194558489 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_001221_470 |
0.1240560742 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 10 |
Hb_002374_510 |
0.1242590606 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 11 |
Hb_000152_570 |
0.1251891552 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 12 |
Hb_000103_300 |
0.1278266974 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 13 |
Hb_000963_120 |
0.1281078691 |
- |
- |
PREDICTED: uncharacterized protein LOC105628103 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005545_110 |
0.1291551781 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 15 |
Hb_000309_070 |
0.1305217845 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 16 |
Hb_000193_070 |
0.1307976776 |
- |
- |
hypothetical protein JCGZ_25373 [Jatropha curcas] |
| 17 |
Hb_007370_010 |
0.1319991017 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 18 |
Hb_006916_140 |
0.1350003703 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 19 |
Hb_141194_020 |
0.1371381556 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 20 |
Hb_001410_080 |
0.1387329685 |
- |
- |
ring finger protein, putative [Ricinus communis] |