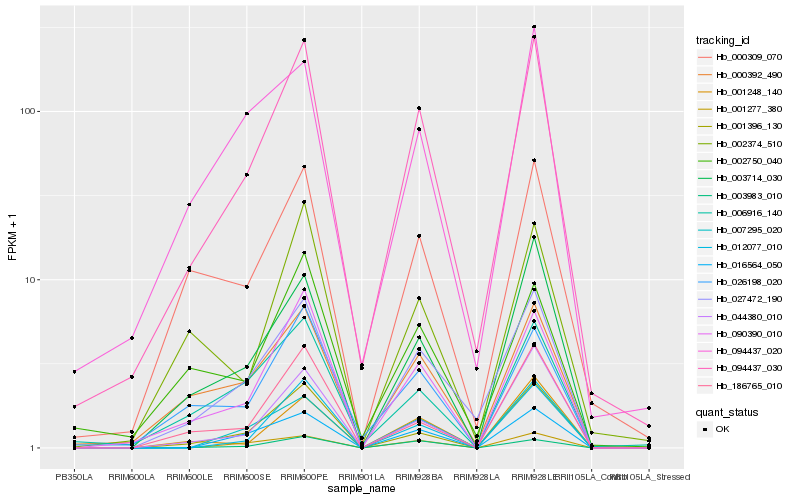
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094437_030 |
0.0 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 2 |
Hb_027472_190 |
0.108449673 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 3 |
Hb_090390_010 |
0.1090132504 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 4 |
Hb_000392_490 |
0.1183608781 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001396_130 |
0.1366410841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000309_070 |
0.139352959 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 7 |
Hb_003714_030 |
0.1405195493 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 8 |
Hb_026198_020 |
0.1417008536 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 9 |
Hb_006916_140 |
0.144556291 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 10 |
Hb_007295_020 |
0.1445709707 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 11 |
Hb_003983_010 |
0.1452827033 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 12 |
Hb_016564_050 |
0.1466462777 |
- |
- |
- |
| 13 |
Hb_012077_010 |
0.1494019978 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 14 |
Hb_044380_010 |
0.1523913386 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 15 |
Hb_001248_140 |
0.1540648268 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_094437_020 |
0.155162358 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002750_040 |
0.1583559361 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 18 |
Hb_186765_010 |
0.1587624744 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 19 |
Hb_002374_510 |
0.1603126154 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 20 |
Hb_001277_380 |
0.1609444155 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Tarenaya hassleriana] |