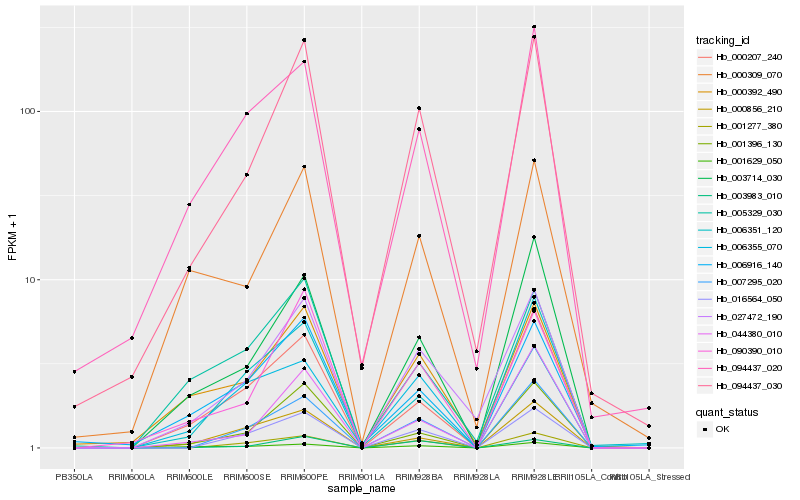
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027472_190 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_094437_030 |
0.108449673 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 3 |
Hb_000392_490 |
0.1434460714 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003714_030 |
0.1511330596 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_016564_050 |
0.1581493037 |
- |
- |
- |
| 6 |
Hb_007295_020 |
0.1585940685 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 7 |
Hb_090390_010 |
0.1587298711 |
- |
- |
hypothetical protein POPTR_0006s28850g [Populus trichocarpa] |
| 8 |
Hb_094437_020 |
0.1617193229 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_001396_130 |
0.1619982232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000207_240 |
0.1632730403 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 11 |
Hb_005329_030 |
0.1638155538 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 12 |
Hb_000309_070 |
0.1644518158 |
- |
- |
PREDICTED: 2-methylene-furan-3-one reductase [Cucumis melo] |
| 13 |
Hb_001277_380 |
0.1670916018 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Tarenaya hassleriana] |
| 14 |
Hb_003983_010 |
0.1760730594 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 15 |
Hb_001629_050 |
0.1777898695 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 16 |
Hb_000856_210 |
0.1779305681 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 17 |
Hb_006916_140 |
0.1790492607 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 18 |
Hb_044380_010 |
0.181040436 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 19 |
Hb_006355_070 |
0.1820218799 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 20 |
Hb_006351_120 |
0.1839055445 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |