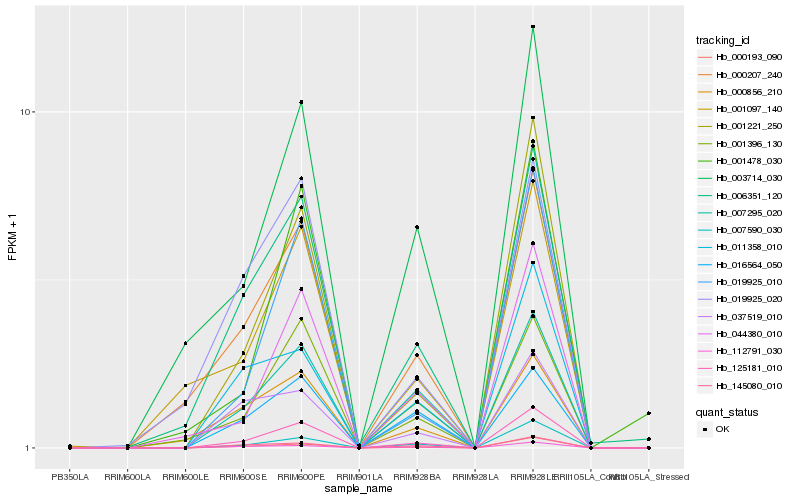
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_125181_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 2 |
Hb_001221_250 |
0.071966222 |
- |
- |
- |
| 3 |
Hb_019925_010 |
0.0954450667 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO4A-like [Jatropha curcas] |
| 4 |
Hb_000193_090 |
0.1005195679 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 5 |
Hb_007590_030 |
0.109651592 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 6 |
Hb_044380_010 |
0.1240141722 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 7 |
Hb_006351_120 |
0.1260979556 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 8 |
Hb_007295_020 |
0.1313058395 |
- |
- |
Uncharacterized protein TCM_034658 [Theobroma cacao] |
| 9 |
Hb_001396_130 |
0.1324563832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_112791_030 |
0.1347344891 |
- |
- |
Uncharacterized protein TCM_027940 [Theobroma cacao] |
| 11 |
Hb_145080_010 |
0.1352953619 |
- |
- |
PREDICTED: uncharacterized protein LOC105643750 [Jatropha curcas] |
| 12 |
Hb_000207_240 |
0.1390577699 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 13 |
Hb_011358_010 |
0.1426313614 |
- |
- |
- |
| 14 |
Hb_003714_030 |
0.1510701458 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 15 |
Hb_037519_010 |
0.1524070987 |
- |
- |
PREDICTED: auxin-induced in root cultures protein 12-like [Jatropha curcas] |
| 16 |
Hb_001478_030 |
0.1616270359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001097_140 |
0.1626843042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000856_210 |
0.1634757628 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 19 |
Hb_016564_050 |
0.1657080007 |
- |
- |
- |
| 20 |
Hb_019925_020 |
0.1661352737 |
- |
- |
electron carrier, putative [Ricinus communis] |