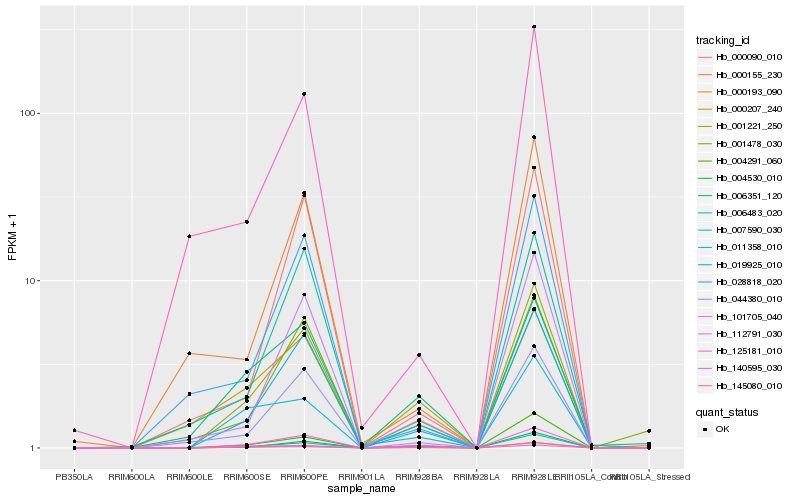
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_250 |
0.0 |
- |
- |
- |
| 2 |
Hb_000193_090 |
0.0707725538 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 3 |
Hb_125181_010 |
0.071966222 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 4 |
Hb_019925_010 |
0.0721146039 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO4A-like [Jatropha curcas] |
| 5 |
Hb_007590_030 |
0.1091786654 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 6 |
Hb_004291_060 |
0.1175732756 |
- |
- |
hypothetical protein F775_42457 [Aegilops tauschii] |
| 7 |
Hb_044380_010 |
0.1474819803 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Malus domestica] |
| 8 |
Hb_001478_030 |
0.1515646511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_028818_020 |
0.1545795847 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 10 |
Hb_101705_040 |
0.1579123804 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000090_010 |
0.1657599898 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 12 |
Hb_000155_230 |
0.1678170474 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 13 |
Hb_004530_010 |
0.1687318255 |
- |
- |
PREDICTED: uncharacterized protein LOC104898079 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_011358_010 |
0.1696654661 |
- |
- |
- |
| 15 |
Hb_140595_030 |
0.1698162346 |
- |
- |
- |
| 16 |
Hb_006351_120 |
0.1699575014 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 17 |
Hb_006483_020 |
0.1723314229 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_112791_030 |
0.1726911916 |
- |
- |
Uncharacterized protein TCM_027940 [Theobroma cacao] |
| 19 |
Hb_145080_010 |
0.1759425143 |
- |
- |
PREDICTED: uncharacterized protein LOC105643750 [Jatropha curcas] |
| 20 |
Hb_000207_240 |
0.1773385359 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |