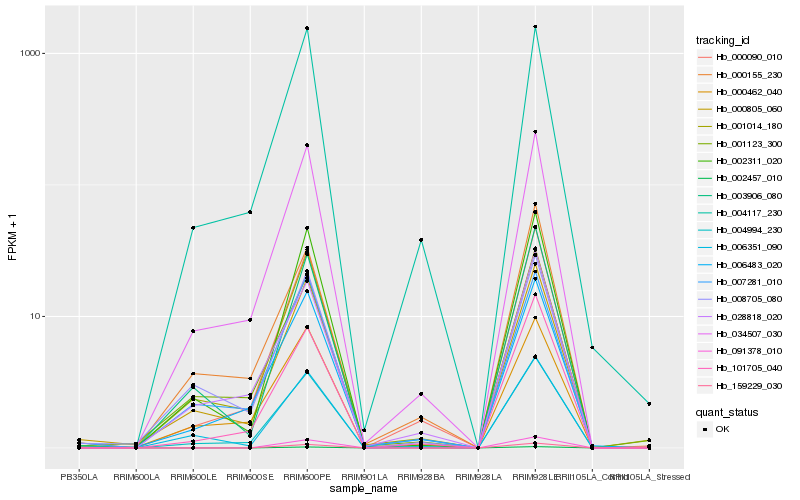
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000090_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 2 |
Hb_004994_230 |
0.041924788 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 3 |
Hb_101705_040 |
0.0555741439 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_034507_030 |
0.0594368604 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 5 |
Hb_006483_020 |
0.0769912271 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002311_020 |
0.0841889355 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 7 |
Hb_028818_020 |
0.0879274858 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 8 |
Hb_004117_230 |
0.0935642492 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 9 |
Hb_000805_060 |
0.1017350972 |
- |
- |
laccase, putative [Ricinus communis] |
| 10 |
Hb_003906_080 |
0.1022254402 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 11 |
Hb_001014_180 |
0.1030235554 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 12 |
Hb_006351_090 |
0.1043537971 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 13 |
Hb_001123_300 |
0.1070875648 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 14 |
Hb_000155_230 |
0.1110199027 |
- |
- |
rac gtpase, putative [Ricinus communis] |
| 15 |
Hb_000462_040 |
0.1125658265 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 16 |
Hb_007281_010 |
0.1152955613 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 17 |
Hb_159229_030 |
0.1155246356 |
- |
- |
hypothetical protein CISIN_1g0034891mg, partial [Citrus sinensis] |
| 18 |
Hb_091378_010 |
0.1156061056 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_008705_080 |
0.115615559 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_002457_010 |
0.1157743596 |
- |
- |
polyprotein [Oryza australiensis] |