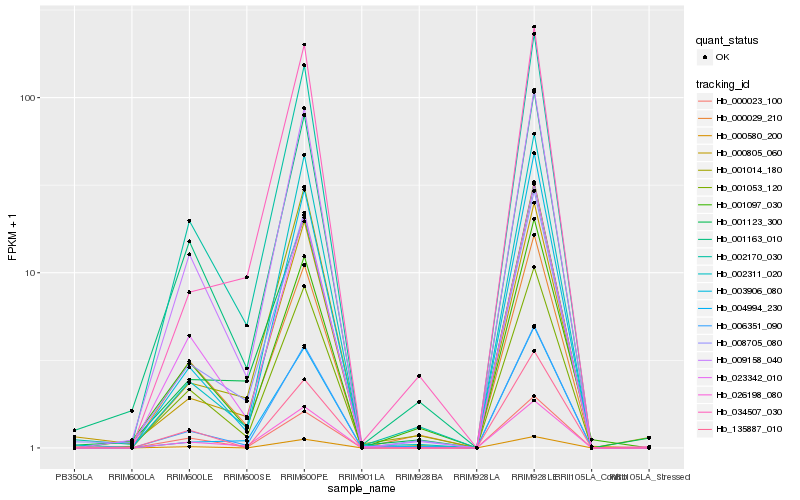
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006351_090 |
0.0 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 2 |
Hb_002170_030 |
0.0447052143 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 3 |
Hb_003906_080 |
0.0531663006 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 4 |
Hb_009158_040 |
0.0565723644 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 5 |
Hb_008705_080 |
0.0630745154 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 6 |
Hb_001053_120 |
0.0639332158 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46-like [Populus euphratica] |
| 7 |
Hb_023342_010 |
0.0687702106 |
- |
- |
- |
| 8 |
Hb_026198_080 |
0.070851972 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 9 |
Hb_002311_020 |
0.0715908658 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 10 |
Hb_000580_200 |
0.0744402222 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000805_060 |
0.0788295415 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_004994_230 |
0.0794774048 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 13 |
Hb_034507_030 |
0.0814894628 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 14 |
Hb_001163_010 |
0.0870125919 |
- |
- |
PREDICTED: syncoilin [Jatropha curcas] |
| 15 |
Hb_001014_180 |
0.0883937057 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 16 |
Hb_000029_210 |
0.0904777107 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_135887_010 |
0.0916938326 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 18 |
Hb_000023_100 |
0.092926911 |
- |
- |
Mechanosensitive channel of small conductance-like 10 [Theobroma cacao] |
| 19 |
Hb_001123_300 |
0.0975254678 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 20 |
Hb_001097_030 |
0.0983609178 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |