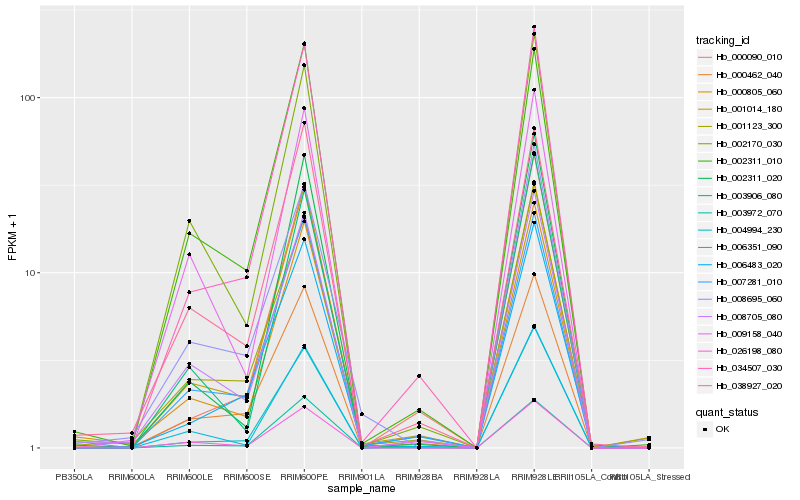
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000805_060 |
0.0 |
- |
- |
laccase, putative [Ricinus communis] |
| 2 |
Hb_008705_080 |
0.0611360519 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14 [Jatropha curcas] |
| 3 |
Hb_002311_020 |
0.0684909566 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 4 |
Hb_034507_030 |
0.0695648252 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 5 |
Hb_004994_230 |
0.0731327495 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 6 |
Hb_007281_010 |
0.0775923898 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 7 |
Hb_001014_180 |
0.0780198211 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 8 |
Hb_006351_090 |
0.0788295415 |
- |
- |
UDP-glycosyltransferase 85A1 [Gossypium arboreum] |
| 9 |
Hb_002170_030 |
0.0825512809 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 10 |
Hb_003906_080 |
0.0833975347 |
- |
- |
Peroxidase 66 precursor, putative [Ricinus communis] |
| 11 |
Hb_026198_080 |
0.0847977503 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Populus euphratica] |
| 12 |
Hb_001123_300 |
0.0850992343 |
- |
- |
hypothetical protein POPTR_0003s16270g [Populus trichocarpa] |
| 13 |
Hb_000462_040 |
0.0877614882 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 14 |
Hb_003972_070 |
0.0886868029 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Citrus sinensis] |
| 15 |
Hb_006483_020 |
0.0903285809 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_038927_020 |
0.0921968006 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 17 |
Hb_009158_040 |
0.0943658488 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |
| 18 |
Hb_002311_010 |
0.0999799626 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 19 |
Hb_000090_010 |
0.1017350972 |
- |
- |
hypothetical protein JCGZ_22567 [Jatropha curcas] |
| 20 |
Hb_008695_060 |
0.1033533353 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |