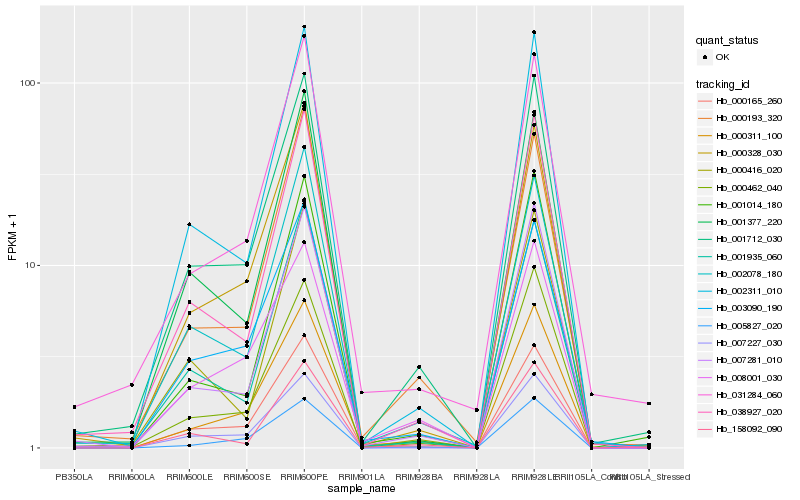
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001712_030 |
0.0 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 2 |
Hb_000462_040 |
0.0574458019 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 3 |
Hb_038927_020 |
0.061231624 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 4 |
Hb_002311_010 |
0.0637149643 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 5 |
Hb_007281_010 |
0.0660900293 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_000311_100 |
0.0664511209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000165_260 |
0.0702800509 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 8 |
Hb_000328_030 |
0.0748810888 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 9 |
Hb_000193_320 |
0.0816877753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002078_180 |
0.081710859 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 8 [Jatropha curcas] |
| 11 |
Hb_007227_030 |
0.0820423667 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 12 |
Hb_031284_060 |
0.0843093683 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 13 |
Hb_001935_060 |
0.0851258067 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 14 |
Hb_008001_030 |
0.0858666451 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003090_190 |
0.0865662066 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 16 |
Hb_158092_090 |
0.0871439005 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001377_220 |
0.087780277 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 18 |
Hb_005827_020 |
0.0878532258 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 19 |
Hb_000416_020 |
0.0878821927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001014_180 |
0.0911608414 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |