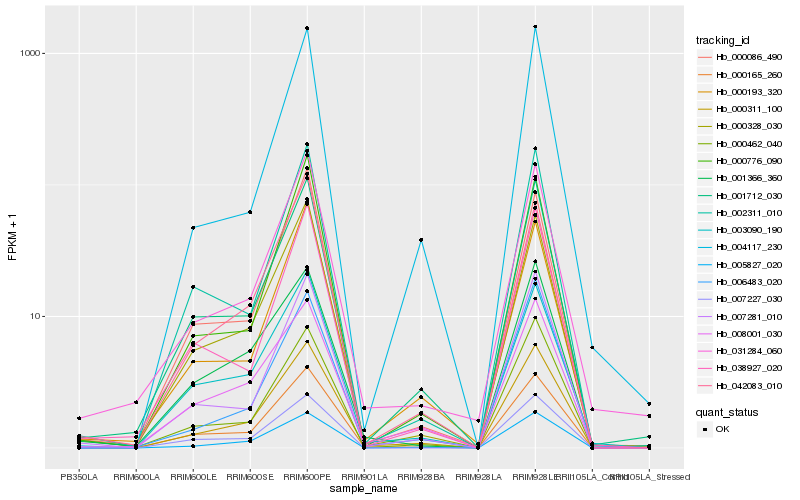
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000311_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005827_020 |
0.0551927996 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 3 |
Hb_000165_260 |
0.0591437768 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 4 |
Hb_001712_030 |
0.0664511209 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 5 |
Hb_000328_030 |
0.0729824754 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 6 |
Hb_008001_030 |
0.0754116368 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003090_190 |
0.0781200957 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 8 |
Hb_000462_040 |
0.0805707591 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 9 |
Hb_007281_010 |
0.0823528956 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_007227_030 |
0.0906513032 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 11 |
Hb_002311_010 |
0.0915251171 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 12 |
Hb_000193_320 |
0.0922741237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_031284_060 |
0.0932553994 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_006483_020 |
0.094049081 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_004117_230 |
0.0967003939 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 16 |
Hb_001366_360 |
0.0972658417 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_000086_490 |
0.0983301441 |
- |
- |
cellulose synthase [Populus tomentosa] |
| 18 |
Hb_038927_020 |
0.0988822548 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC13 [Cucumis melo] |
| 19 |
Hb_042083_010 |
0.1007607734 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 20 |
Hb_000776_090 |
0.1031030527 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |