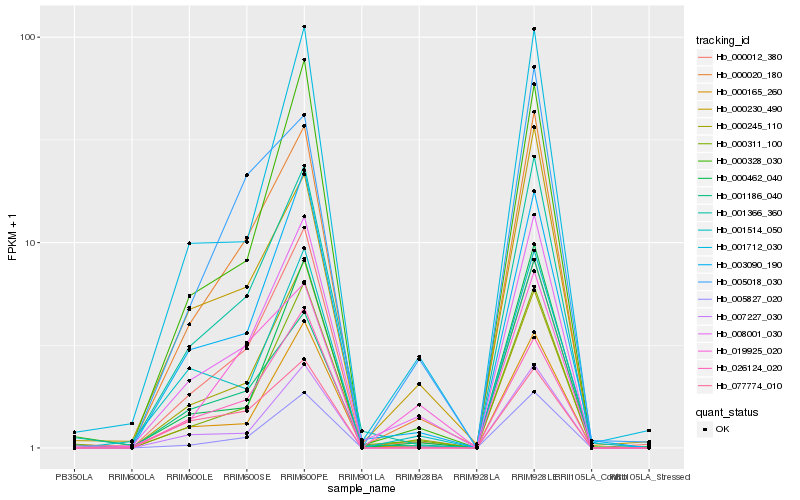
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_180 |
0.0 |
- |
- |
laccase, putative [Ricinus communis] |
| 2 |
Hb_001366_360 |
0.0678661085 |
- |
- |
catalytic, putative [Ricinus communis] |
| 3 |
Hb_008001_030 |
0.078815391 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005827_020 |
0.0842584271 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 5 |
Hb_005018_030 |
0.1055731609 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 6 |
Hb_007227_030 |
0.1071280721 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 7 |
Hb_000311_100 |
0.1104978149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000230_490 |
0.1149437808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003090_190 |
0.1186050057 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 10 |
Hb_000165_260 |
0.1193739137 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 11 |
Hb_000012_380 |
0.1195543713 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001712_030 |
0.1236446345 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 19 [Jatropha curcas] |
| 13 |
Hb_000245_110 |
0.1242859796 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 14 |
Hb_001186_040 |
0.1255876949 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 15 |
Hb_000462_040 |
0.1270441836 |
- |
- |
ATSMC2, putative [Ricinus communis] |
| 16 |
Hb_000328_030 |
0.129144908 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 17 |
Hb_077774_010 |
0.1343530499 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 18 |
Hb_026124_020 |
0.1355282943 |
- |
- |
PREDICTED: uncharacterized protein LOC105644241 isoform X1 [Jatropha curcas] |
| 19 |
Hb_019925_020 |
0.1358039677 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 20 |
Hb_001514_050 |
0.1369984818 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |