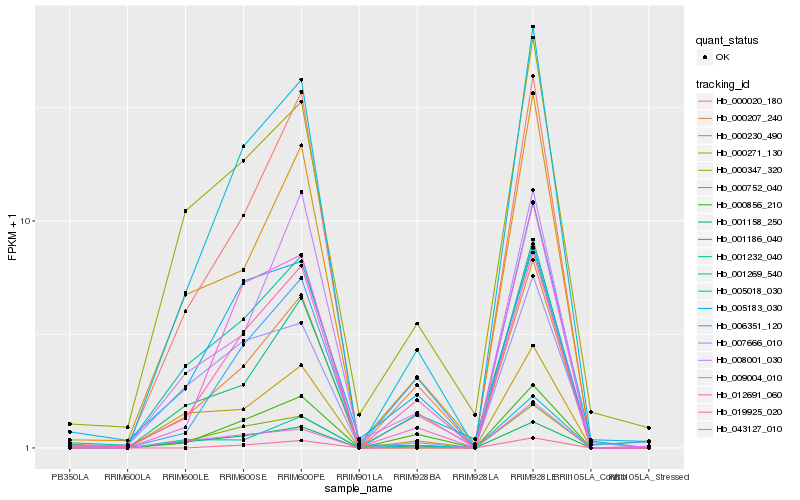
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005018_030 |
0.0 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 2 |
Hb_012691_060 |
0.0829419439 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like isoform X1 [Prunus mume] |
| 3 |
Hb_000020_180 |
0.1055731609 |
- |
- |
laccase, putative [Ricinus communis] |
| 4 |
Hb_019925_020 |
0.1140571416 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 5 |
Hb_000230_490 |
0.1188934057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000752_040 |
0.1220137679 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_005183_030 |
0.12708984 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 8 |
Hb_000347_320 |
0.1320990971 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 9 |
Hb_001186_040 |
0.1363762653 |
- |
- |
Peroxidase 3 precursor, putative [Ricinus communis] |
| 10 |
Hb_001158_250 |
0.1389441265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001269_540 |
0.140013974 |
- |
- |
hypothetical protein VITISV_038016 [Vitis vinifera] |
| 12 |
Hb_007666_010 |
0.140781269 |
- |
- |
PREDICTED: uncharacterized protein LOC105649266 [Jatropha curcas] |
| 13 |
Hb_000856_210 |
0.1430971304 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 14 |
Hb_000207_240 |
0.1454849907 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 15 |
Hb_006351_120 |
0.1508423028 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 16 |
Hb_009004_010 |
0.1510629295 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 17 |
Hb_008001_030 |
0.1516183385 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_043127_010 |
0.1521723359 |
- |
- |
PREDICTED: uncharacterized protein LOC105648822 [Jatropha curcas] |
| 19 |
Hb_000271_130 |
0.1531706551 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 20 |
Hb_001232_040 |
0.1533965596 |
- |
- |
PREDICTED: uncharacterized protein LOC105634974 [Jatropha curcas] |