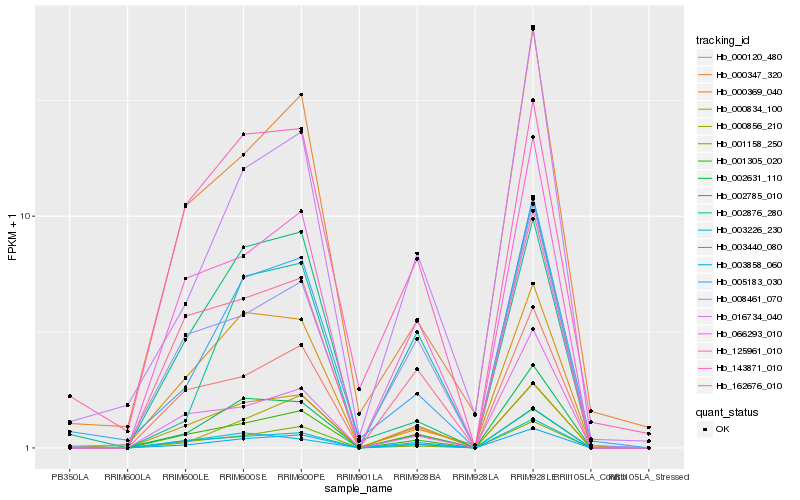
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_110 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 2 |
Hb_005183_030 |
0.1010578741 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 3 |
Hb_003226_230 |
0.1112105408 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 4 |
Hb_003858_060 |
0.1216453717 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 5 |
Hb_000369_040 |
0.1301757766 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 6 |
Hb_125961_010 |
0.1326352172 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_162676_010 |
0.1334690135 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000834_100 |
0.1355057421 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 9 |
Hb_002785_010 |
0.1374520325 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 10 |
Hb_000120_480 |
0.1387122248 |
- |
- |
- |
| 11 |
Hb_001158_250 |
0.1410454392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002876_280 |
0.1417783269 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 13 |
Hb_016734_040 |
0.1423187524 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 14 |
Hb_008461_070 |
0.1443730153 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003440_080 |
0.1489125748 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 16 |
Hb_000347_320 |
0.1516645421 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_000856_210 |
0.1520106528 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_066293_010 |
0.1530016779 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 19 |
Hb_143871_010 |
0.1537626401 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_001305_020 |
0.1539666296 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |