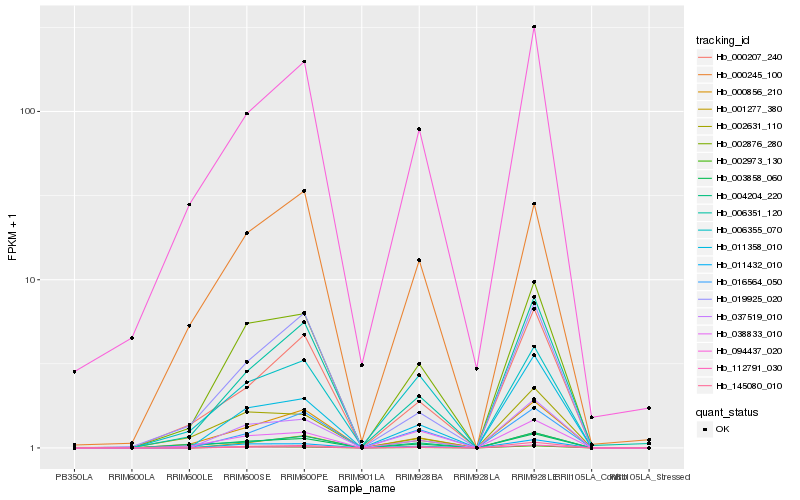
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_280 |
0.0 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 2 |
Hb_011432_010 |
0.0943314841 |
- |
- |
hypothetical protein JCGZ_23781 [Jatropha curcas] |
| 3 |
Hb_003858_060 |
0.1012258503 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 4 |
Hb_000856_210 |
0.1014522607 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 5 |
Hb_002973_130 |
0.1150318442 |
- |
- |
PREDICTED: uncharacterized protein LOC104609908 [Nelumbo nucifera] |
| 6 |
Hb_037519_010 |
0.1211800008 |
- |
- |
PREDICTED: auxin-induced in root cultures protein 12-like [Jatropha curcas] |
| 7 |
Hb_145080_010 |
0.1331516171 |
- |
- |
PREDICTED: uncharacterized protein LOC105643750 [Jatropha curcas] |
| 8 |
Hb_006355_070 |
0.1356735405 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 9 |
Hb_006351_120 |
0.1357452642 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 10 |
Hb_094437_020 |
0.1359590401 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_112791_030 |
0.13805612 |
- |
- |
Uncharacterized protein TCM_027940 [Theobroma cacao] |
| 12 |
Hb_011358_010 |
0.1410285728 |
- |
- |
- |
| 13 |
Hb_002631_110 |
0.1417783269 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_000207_240 |
0.1429690849 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 15 |
Hb_019925_020 |
0.1472160208 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 16 |
Hb_038833_010 |
0.1506578279 |
- |
- |
polyprotein [Ananas comosus] |
| 17 |
Hb_000245_100 |
0.15183004 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 18 |
Hb_004204_220 |
0.1619952958 |
- |
- |
- |
| 19 |
Hb_016564_050 |
0.1630701651 |
- |
- |
- |
| 20 |
Hb_001277_380 |
0.1634862067 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 3 [Tarenaya hassleriana] |