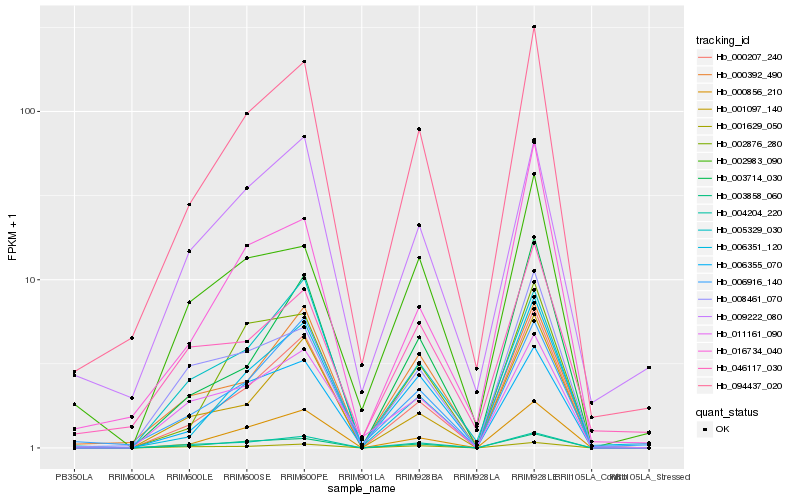
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094437_020 |
0.0 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_003858_060 |
0.1096804581 |
- |
- |
Germin-like protein subfamily 1 member 18 [Theobroma cacao] |
| 3 |
Hb_000207_240 |
0.1098231595 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 4 |
Hb_006916_140 |
0.1146862948 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 5 |
Hb_000856_210 |
0.1165192864 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 6 |
Hb_000392_490 |
0.1210485533 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004204_220 |
0.1217620398 |
- |
- |
- |
| 8 |
Hb_006351_120 |
0.1232702049 |
- |
- |
hypothetical protein CICLE_v10030803mg [Citrus clementina] |
| 9 |
Hb_046117_030 |
0.1340152948 |
- |
- |
sialyltransferase, putative [Ricinus communis] |
| 10 |
Hb_002876_280 |
0.1359590401 |
- |
- |
hypothetical protein RCOM_0686540 [Ricinus communis] |
| 11 |
Hb_008461_070 |
0.1365997379 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_016734_040 |
0.1366219499 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 13 |
Hb_009222_080 |
0.1371756997 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 14 |
Hb_001629_050 |
0.1378031443 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 15 |
Hb_011161_090 |
0.1384670355 |
- |
- |
- |
| 16 |
Hb_003714_030 |
0.1390864522 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 17 |
Hb_002983_090 |
0.1424328329 |
- |
- |
Altered inheritance of mitochondria protein 32 [Glycine soja] |
| 18 |
Hb_005329_030 |
0.1433812447 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 19 |
Hb_006355_070 |
0.1457476759 |
transcription factor |
TF Family: bHLH |
Phytochrome-interacting factor, putative [Ricinus communis] |
| 20 |
Hb_001097_140 |
0.1459710567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |