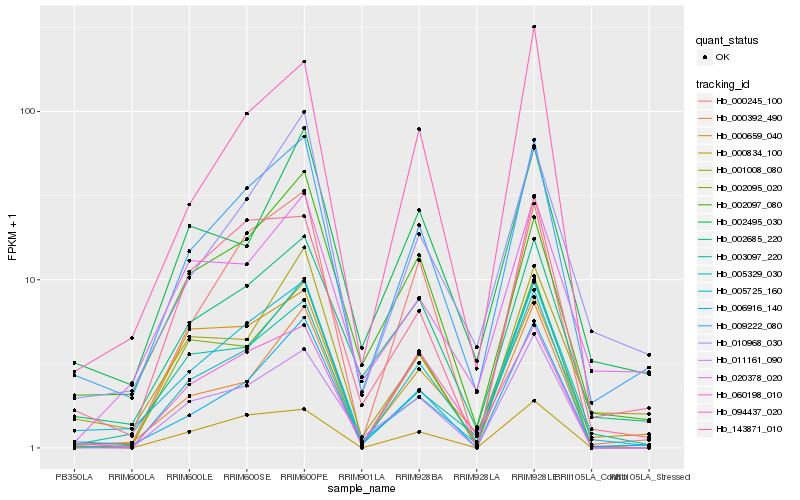
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_080 |
0.0 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 2 |
Hb_002685_220 |
0.0999609458 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006916_140 |
0.105846068 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 4 |
Hb_005725_160 |
0.1162389207 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 5 |
Hb_010968_030 |
0.1231409678 |
- |
- |
PREDICTED: uncharacterized protein LOC105643149 [Jatropha curcas] |
| 6 |
Hb_005329_030 |
0.1259021915 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 7 |
Hb_000245_100 |
0.1268707866 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 8 |
Hb_002097_080 |
0.1327302556 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 9 |
Hb_001008_080 |
0.1336142703 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 10 |
Hb_094437_020 |
0.1371756997 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_003097_220 |
0.1413942109 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 12 |
Hb_002495_030 |
0.1444984552 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002095_020 |
0.1449379665 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000392_490 |
0.146270346 |
- |
- |
PREDICTED: uncharacterized protein LOC105640495 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000659_040 |
0.1477955462 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_143871_010 |
0.1481212073 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 17 |
Hb_060198_010 |
0.1484056913 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 18 |
Hb_020378_020 |
0.1492168153 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000834_100 |
0.150185595 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 20 |
Hb_011161_090 |
0.1514678834 |
- |
- |
- |