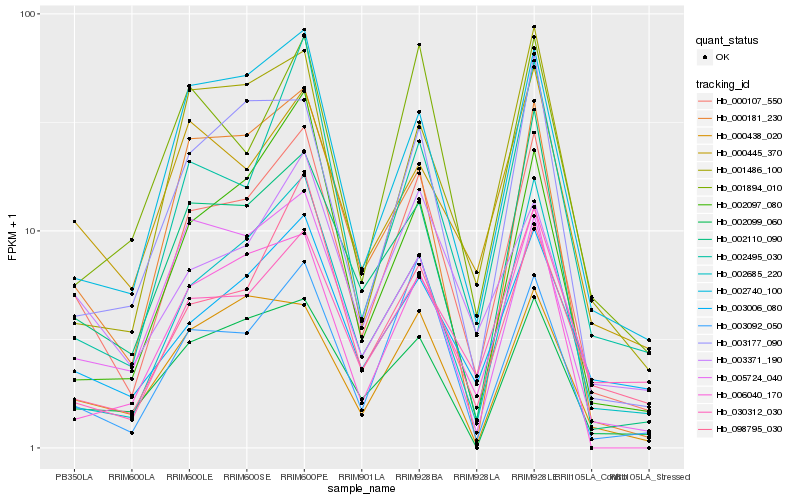
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_550 |
0.0 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 2 |
Hb_002740_100 |
0.1171267609 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 3 |
Hb_002685_220 |
0.1178350243 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002110_090 |
0.1279216838 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 5 |
Hb_003092_050 |
0.1289945121 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 6 |
Hb_000181_230 |
0.132339205 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_003006_080 |
0.1349946115 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002099_060 |
0.1402819993 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 9 |
Hb_003371_190 |
0.1409861575 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 10 |
Hb_001894_010 |
0.1426717986 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 11 |
Hb_005724_040 |
0.1433436312 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 12 |
Hb_003177_090 |
0.143637165 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 13 |
Hb_098795_030 |
0.143655779 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 14 |
Hb_001486_100 |
0.1441568198 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 15 |
Hb_002495_030 |
0.1452389238 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 16 |
Hb_030312_030 |
0.145705897 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000445_370 |
0.1459886328 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 18 |
Hb_000438_020 |
0.1480464258 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 19 |
Hb_002097_080 |
0.1487830204 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 20 |
Hb_006040_170 |
0.1498699094 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |