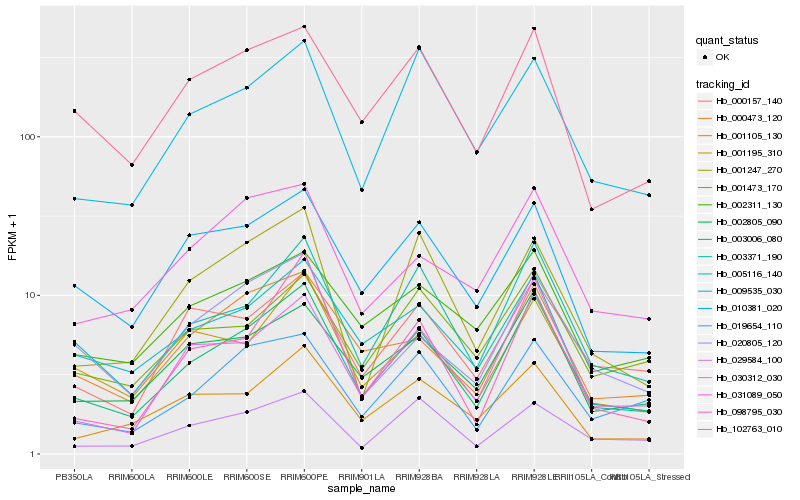
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003006_080 |
0.0 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_030312_030 |
0.1049710086 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005116_140 |
0.1067595451 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 4 |
Hb_002805_090 |
0.1080373679 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_000473_120 |
0.1124241782 |
- |
- |
hypothetical protein JCGZ_16177 [Jatropha curcas] |
| 6 |
Hb_019654_110 |
0.1130072458 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 7 |
Hb_010381_020 |
0.1141050439 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 8 |
Hb_001195_310 |
0.1145520594 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001105_130 |
0.1148523811 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_102763_010 |
0.1187171427 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 11 |
Hb_000157_140 |
0.1198557863 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_009535_030 |
0.1206894796 |
- |
- |
CP2 [Hevea brasiliensis] |
| 13 |
Hb_020805_120 |
0.122476448 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 14 |
Hb_031089_050 |
0.1230572698 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_002311_130 |
0.124842652 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_098795_030 |
0.1258161374 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 17 |
Hb_003371_190 |
0.1261066512 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 18 |
Hb_029584_100 |
0.1264467545 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 19 |
Hb_001247_270 |
0.1271853324 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001473_170 |
0.1285504133 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |