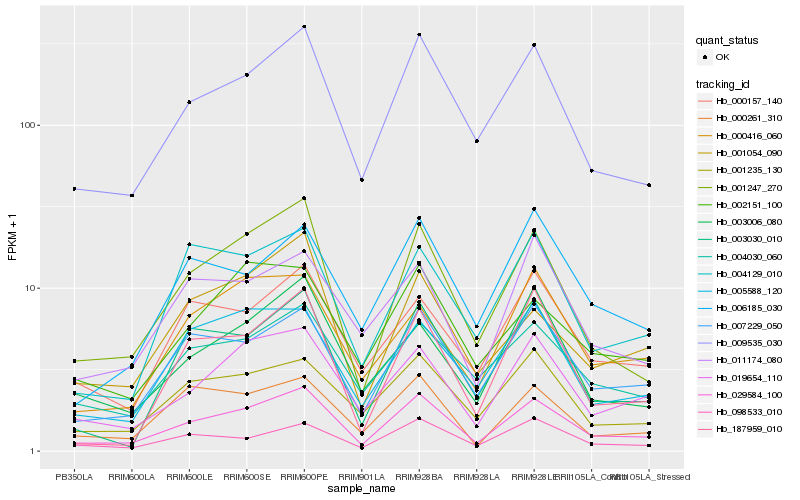
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029584_100 |
0.0 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 2 |
Hb_009535_030 |
0.0851590989 |
- |
- |
CP2 [Hevea brasiliensis] |
| 3 |
Hb_001247_270 |
0.0883564898 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007229_050 |
0.0963102436 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_019654_110 |
0.105702068 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |
| 6 |
Hb_001235_130 |
0.1212598091 |
- |
- |
- |
| 7 |
Hb_098533_010 |
0.1218680813 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 8 |
Hb_003030_010 |
0.1225498765 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 9 |
Hb_004030_060 |
0.1244734093 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 10 |
Hb_003006_080 |
0.1264467545 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000261_310 |
0.1275967337 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 12 |
Hb_000157_140 |
0.1292440128 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_005588_120 |
0.1325282028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001054_090 |
0.1330153715 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 15 |
Hb_004129_010 |
0.1338880221 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 16 |
Hb_000416_060 |
0.1353335532 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 17 |
Hb_002151_100 |
0.1357431817 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 18 |
Hb_011174_080 |
0.1361985509 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 19 |
Hb_187959_010 |
0.1386479116 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 20 |
Hb_006185_030 |
0.1387503318 |
- |
- |
threonine synthase, putative [Ricinus communis] |