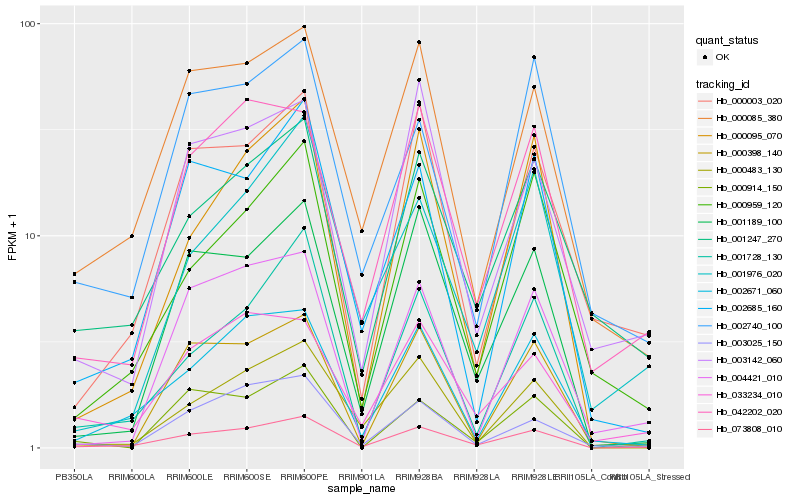
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073808_010 |
0.0 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_001189_100 |
0.1152865102 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2-like [Jatropha curcas] |
| 3 |
Hb_002671_060 |
0.1198410482 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 4 |
Hb_000085_380 |
0.1209697153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001728_130 |
0.1250317132 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001247_270 |
0.1329394677 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_042202_020 |
0.1335304998 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 8 |
Hb_002685_160 |
0.1341593257 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000483_130 |
0.136425044 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000959_120 |
0.1368462301 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 11 |
Hb_000003_020 |
0.1385753479 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 12 |
Hb_033234_010 |
0.1390026929 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 13 |
Hb_000095_070 |
0.1410204898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003025_150 |
0.1426734259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000398_140 |
0.1452440193 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 16 |
Hb_002740_100 |
0.1522158813 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_003142_060 |
0.1527389281 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000914_150 |
0.1540674086 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 19 |
Hb_004421_010 |
0.1545608116 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 20 |
Hb_001976_020 |
0.1548991524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |