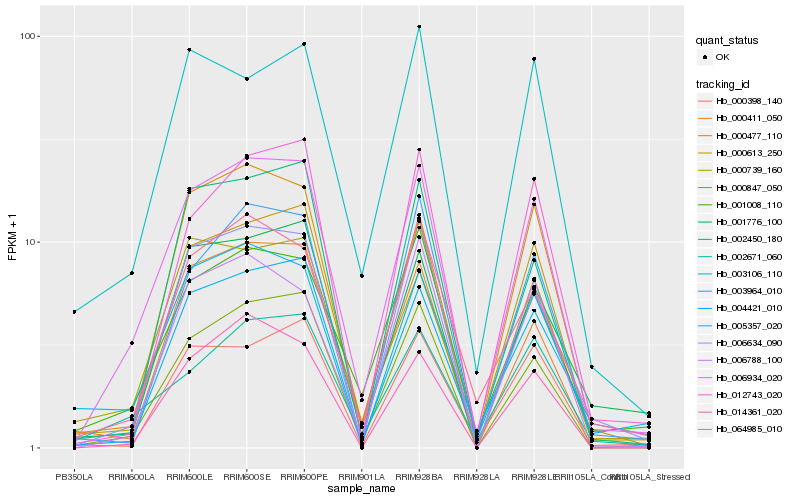
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006934_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s09051g [Populus trichocarpa] |
| 2 |
Hb_000847_050 |
0.1031194969 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 3 |
Hb_000739_160 |
0.1071086821 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_002671_060 |
0.107848452 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 5 |
Hb_006634_090 |
0.1086868997 |
- |
- |
PREDICTED: uncharacterized protein LOC105646581 [Jatropha curcas] |
| 6 |
Hb_001008_110 |
0.1117931808 |
- |
- |
- |
| 7 |
Hb_004421_010 |
0.1144705647 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_000477_110 |
0.1185440295 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |
| 9 |
Hb_064985_010 |
0.1196127835 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 10 |
Hb_012743_020 |
0.1257735957 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000398_140 |
0.1264337771 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 12 |
Hb_000411_050 |
0.1266993336 |
- |
- |
hypothetical protein AMTR_s00066p00166230 [Amborella trichopoda] |
| 13 |
Hb_003106_110 |
0.1301791065 |
- |
- |
betaine aldehyde dehydrogenase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001776_100 |
0.130573153 |
- |
- |
PREDICTED: outer envelope pore protein 16, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005357_020 |
0.1307872088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003964_010 |
0.1309026503 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0008s09360g [Populus trichocarpa] |
| 17 |
Hb_000613_250 |
0.1326705733 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 18 |
Hb_006788_100 |
0.1332838367 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002450_180 |
0.1342436331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_014361_020 |
0.1346173452 |
- |
- |
PREDICTED: probable protein phosphatase 2C 78 [Jatropha curcas] |