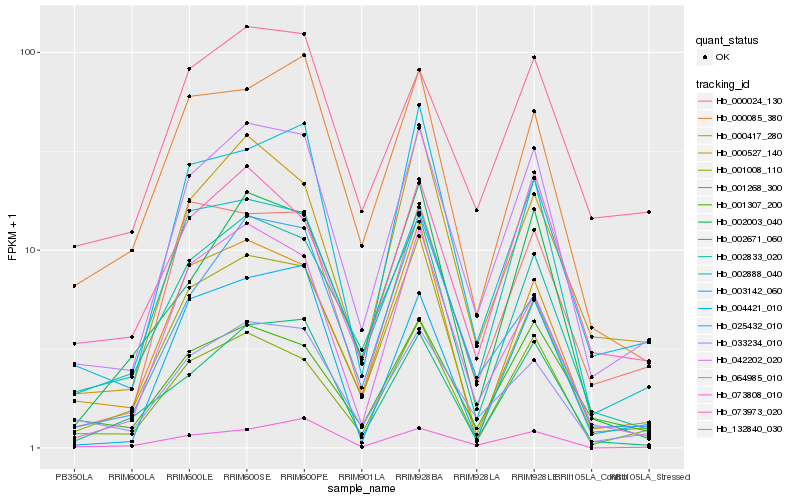
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042202_020 |
0.0 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 2 |
Hb_033234_010 |
0.0894138748 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 3 |
Hb_001268_300 |
0.105789388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001008_110 |
0.1099623586 |
- |
- |
- |
| 5 |
Hb_002833_020 |
0.1104058946 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_000527_140 |
0.1125512094 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 7 |
Hb_003142_060 |
0.1163760755 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002888_040 |
0.1169246404 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 9 |
Hb_001307_200 |
0.1172496384 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 10 |
Hb_000024_130 |
0.1201120195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002671_060 |
0.1226714982 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 12 |
Hb_132840_030 |
0.1228807319 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 13 |
Hb_025432_010 |
0.1235954428 |
- |
- |
hypothetical protein JCGZ_21627 [Jatropha curcas] |
| 14 |
Hb_064985_010 |
0.1246850738 |
- |
- |
Leucine-rich repeat transmembrane protein kinase, putative [Theobroma cacao] |
| 15 |
Hb_000417_280 |
0.1268762278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000085_380 |
0.1269188825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_073973_020 |
0.1284825744 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 18 |
Hb_002003_040 |
0.1316738196 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 19 |
Hb_073808_010 |
0.1335304998 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_004421_010 |
0.1355918698 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |