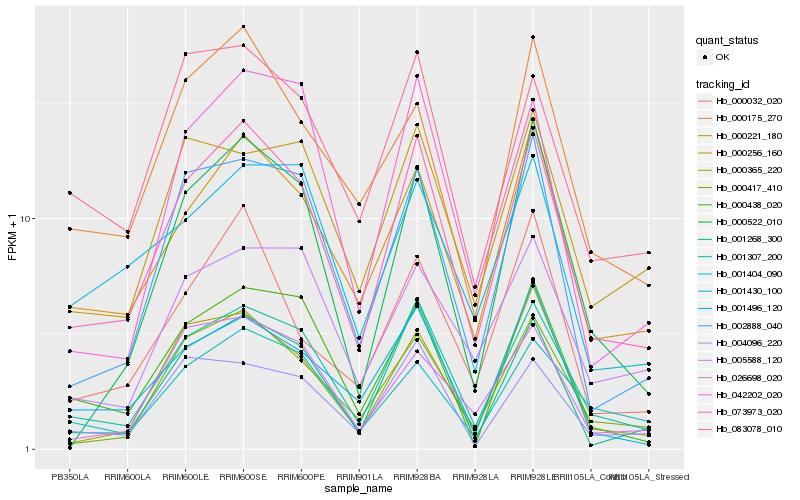
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073973_020 |
0.0 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 2 |
Hb_001307_200 |
0.0561109282 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 3 |
Hb_000221_180 |
0.0902648215 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 4 |
Hb_000417_410 |
0.1047382623 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001268_300 |
0.1075717349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002888_040 |
0.1136924408 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 7 |
Hb_001496_120 |
0.1221351206 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_001404_090 |
0.1231849588 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 9 |
Hb_004096_220 |
0.1236162237 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 10 |
Hb_083078_010 |
0.1243014026 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 11 |
Hb_000365_220 |
0.1246049362 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 12 |
Hb_000175_270 |
0.1256322367 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000438_020 |
0.1274960885 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 14 |
Hb_042202_020 |
0.1284825744 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 15 |
Hb_026698_020 |
0.1292160981 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_000032_020 |
0.1317025226 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |
| 17 |
Hb_005588_120 |
0.1318822333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000256_160 |
0.136380343 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 19 |
Hb_000522_010 |
0.1364345151 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 20 |
Hb_001430_100 |
0.1368415406 |
- |
- |
cytochrome P450, putative [Ricinus communis] |