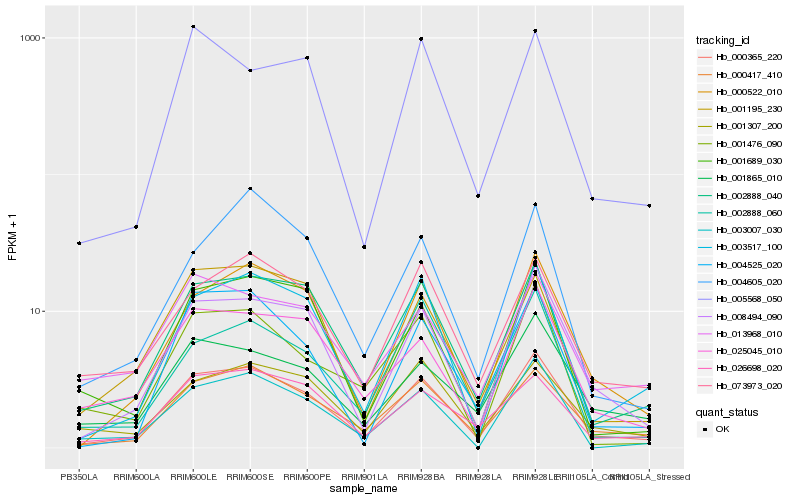
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_220 |
0.0 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 2 |
Hb_008494_090 |
0.0850926834 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 3 |
Hb_000522_010 |
0.0904695033 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 4 |
Hb_000417_410 |
0.0915744597 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002888_040 |
0.0939060546 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 6 |
Hb_003517_100 |
0.1070828905 |
transcription factor |
TF Family: GRAS |
- |
| 7 |
Hb_004525_020 |
0.1245674881 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 8 |
Hb_073973_020 |
0.1246049362 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 9 |
Hb_026698_020 |
0.1247778331 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 10 |
Hb_025045_010 |
0.1248823119 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_001865_010 |
0.1274787529 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 12 |
Hb_001689_030 |
0.1313416217 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 13 |
Hb_003007_030 |
0.1323022763 |
- |
- |
- |
| 14 |
Hb_001307_200 |
0.136289047 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 15 |
Hb_001195_230 |
0.1377896881 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 16 |
Hb_002888_060 |
0.143183948 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005568_050 |
0.1455079691 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 18 |
Hb_001476_090 |
0.1463192376 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 19 |
Hb_004605_020 |
0.1469371135 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 20 |
Hb_013968_010 |
0.1491566513 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |