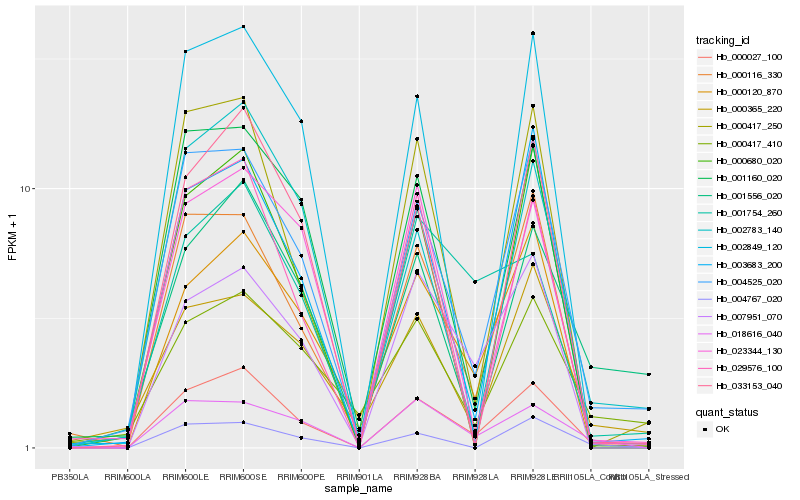
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_100 |
0.0 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 2 |
Hb_004525_020 |
0.1048824611 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 3 |
Hb_018616_040 |
0.150029086 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000116_330 |
0.1501088737 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_000417_250 |
0.1575978047 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 6 |
Hb_001556_020 |
0.1578198777 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_007951_070 |
0.1611436152 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 8 |
Hb_003683_200 |
0.1622768552 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_000120_870 |
0.1640544144 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 10 |
Hb_002783_140 |
0.1676655133 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 11 |
Hb_029576_100 |
0.1681728309 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 12 |
Hb_001754_260 |
0.1702006329 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000417_410 |
0.177294161 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001160_020 |
0.1782330252 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 15 |
Hb_000365_220 |
0.1786270008 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 16 |
Hb_033153_040 |
0.1788084229 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000680_020 |
0.1803763912 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002849_120 |
0.1813847418 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 19 |
Hb_004767_020 |
0.1818763479 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 20 |
Hb_023344_130 |
0.185347067 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |