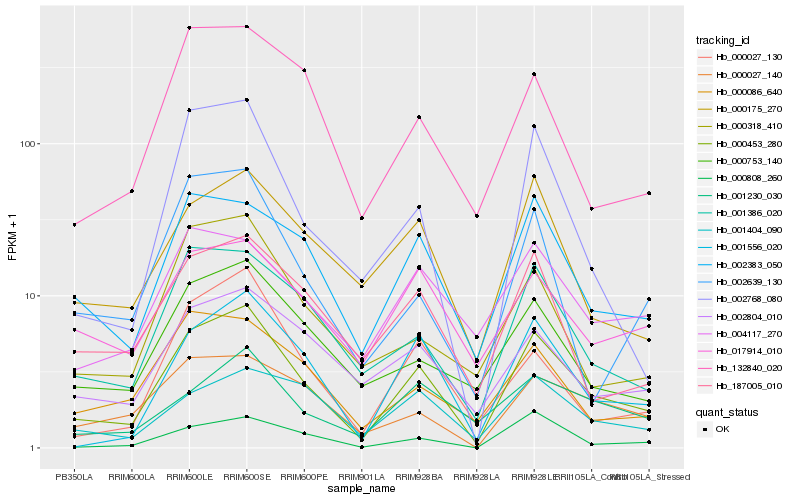
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_280 |
0.0 |
- |
- |
negative cofactor 2 transcriptional co-repressor, putative [Ricinus communis] |
| 2 |
Hb_001556_020 |
0.1445178078 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 3 |
Hb_001386_020 |
0.150045556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002383_050 |
0.153335897 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 5 |
Hb_000175_270 |
0.165865566 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000753_140 |
0.1680920988 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 7 |
Hb_000808_260 |
0.1686712205 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 8 |
Hb_187005_010 |
0.1694092295 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 9 |
Hb_002768_080 |
0.1703679203 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl-diphosphate synthase [Hevea brasiliensis] |
| 10 |
Hb_002804_010 |
0.1737736279 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 11 |
Hb_000027_140 |
0.1756605929 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 12 |
Hb_000086_640 |
0.1763278439 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_001404_090 |
0.1777052579 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 14 |
Hb_132840_020 |
0.1805885129 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 15 |
Hb_002639_130 |
0.1820728948 |
- |
- |
hypothetical protein JCGZ_08129 [Jatropha curcas] |
| 16 |
Hb_000027_130 |
0.1827098389 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 17 |
Hb_000318_410 |
0.1840078222 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 18 |
Hb_017914_010 |
0.1842697992 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SUPPRESSOR OF FRI 4 [Jatropha curcas] |
| 19 |
Hb_004117_270 |
0.18483304 |
- |
- |
- |
| 20 |
Hb_001230_030 |
0.1849815735 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |