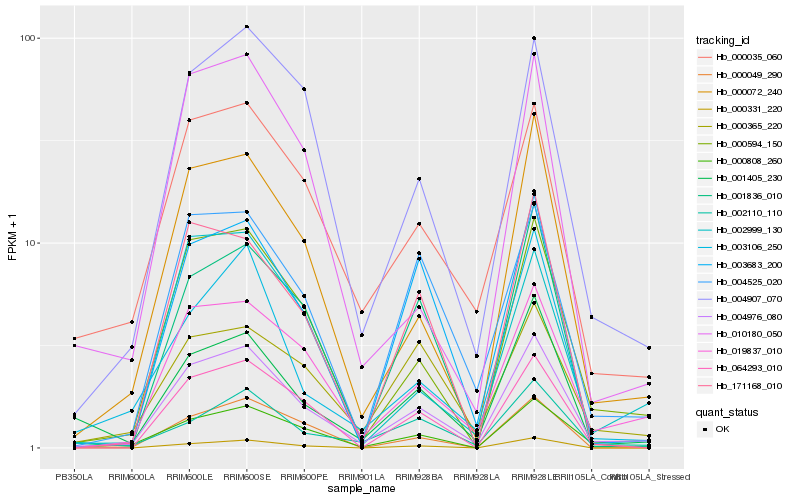
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004976_080 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 2 |
Hb_019837_010 |
0.0969699561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000594_150 |
0.0993701357 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 4 |
Hb_000072_240 |
0.1071276821 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_004907_070 |
0.120299082 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 6 |
Hb_000035_060 |
0.131456229 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 7 |
Hb_004525_020 |
0.1319266042 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 8 |
Hb_000808_260 |
0.1319880564 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 9 |
Hb_064293_010 |
0.13841967 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_003683_200 |
0.1430304017 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 11 |
Hb_001405_230 |
0.1432845206 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 12 |
Hb_001836_010 |
0.15285943 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 13 |
Hb_171168_010 |
0.1529023094 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000365_220 |
0.152978841 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 15 |
Hb_000331_220 |
0.1545554586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010180_050 |
0.156579929 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_003106_250 |
0.1585873131 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 18 |
Hb_002999_130 |
0.1589068147 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 19 |
Hb_000049_290 |
0.1590005572 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 20 |
Hb_002110_110 |
0.1607121429 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |