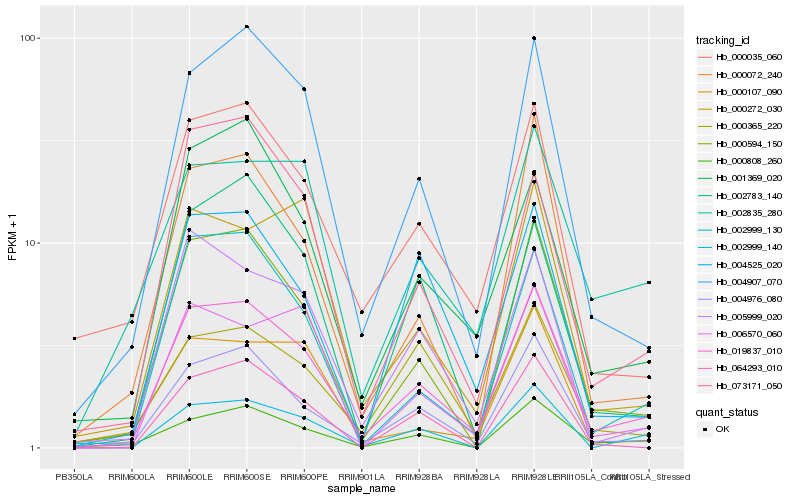
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019837_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004976_080 |
0.0969699561 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 3 |
Hb_000594_150 |
0.107455429 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 4 |
Hb_004907_070 |
0.1133391132 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 5 |
Hb_000808_260 |
0.1321760818 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 6 |
Hb_000072_240 |
0.1343039354 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002999_130 |
0.1352995702 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 8 |
Hb_002999_140 |
0.137433213 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 9 |
Hb_000035_060 |
0.1380955205 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 10 |
Hb_005999_020 |
0.1391615231 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 11 |
Hb_004525_020 |
0.1486959113 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 12 |
Hb_001369_020 |
0.1515229328 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 13 |
Hb_002835_280 |
0.1550743824 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 14 |
Hb_006570_060 |
0.157599821 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 15 |
Hb_073171_050 |
0.1576841245 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 16 |
Hb_000272_030 |
0.1612294462 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 17 |
Hb_064293_010 |
0.1624493403 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_002783_140 |
0.1636701492 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 19 |
Hb_000107_090 |
0.1656694365 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 20 |
Hb_000365_220 |
0.1660318217 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |