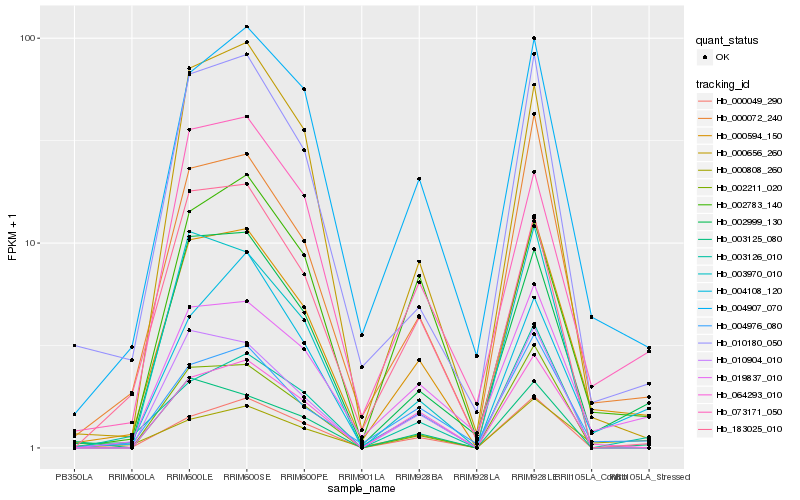
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_150 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 2 |
Hb_000072_240 |
0.0879462157 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_004976_080 |
0.0993701357 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 4 |
Hb_019837_010 |
0.107455429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002999_130 |
0.1085265283 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 6 |
Hb_004907_070 |
0.116928016 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 7 |
Hb_010180_050 |
0.1176770844 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 8 |
Hb_064293_010 |
0.1293969183 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 9 |
Hb_000808_260 |
0.1342358742 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 10 |
Hb_002211_020 |
0.1406137514 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 11 |
Hb_010904_010 |
0.1420359098 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000049_290 |
0.144494063 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_073171_050 |
0.1529265151 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 14 |
Hb_003970_010 |
0.1535500541 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_000656_260 |
0.1542257776 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 16 |
Hb_003125_080 |
0.1549437146 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 17 |
Hb_183025_010 |
0.1570308403 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 18 |
Hb_002783_140 |
0.1583528592 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 19 |
Hb_003126_010 |
0.1600458671 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 20 |
Hb_004108_120 |
0.1601994687 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |