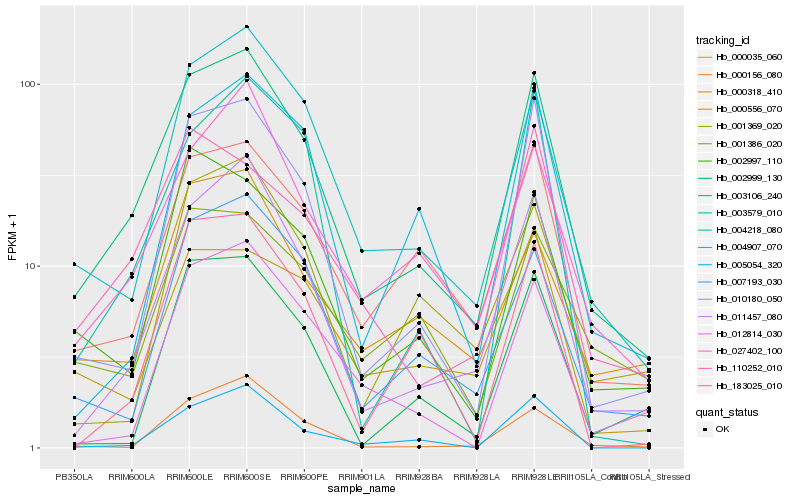
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_240 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 2 |
Hb_011457_080 |
0.1102046226 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004218_080 |
0.1273093876 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 4 |
Hb_010180_050 |
0.1297871113 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 5 |
Hb_027402_100 |
0.1323211161 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 6 |
Hb_000035_060 |
0.1348695535 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Jatropha curcas] |
| 7 |
Hb_007193_030 |
0.1382296408 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000318_410 |
0.1588844783 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 9 |
Hb_005054_320 |
0.159328522 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 10 |
Hb_002997_110 |
0.1600884257 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 11 |
Hb_110252_010 |
0.1624768671 |
- |
- |
hypothetical protein JCGZ_16705 [Jatropha curcas] |
| 12 |
Hb_004907_070 |
0.1635198354 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_000156_080 |
0.1647235808 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 14 |
Hb_012814_030 |
0.1653447711 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 15 |
Hb_003579_010 |
0.166085963 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 16 |
Hb_001369_020 |
0.167992356 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 17 |
Hb_001386_020 |
0.1683301731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000556_070 |
0.1696757977 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At2g45590 [Jatropha curcas] |
| 19 |
Hb_183025_010 |
0.16994898 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 20 |
Hb_002999_130 |
0.1704846189 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |