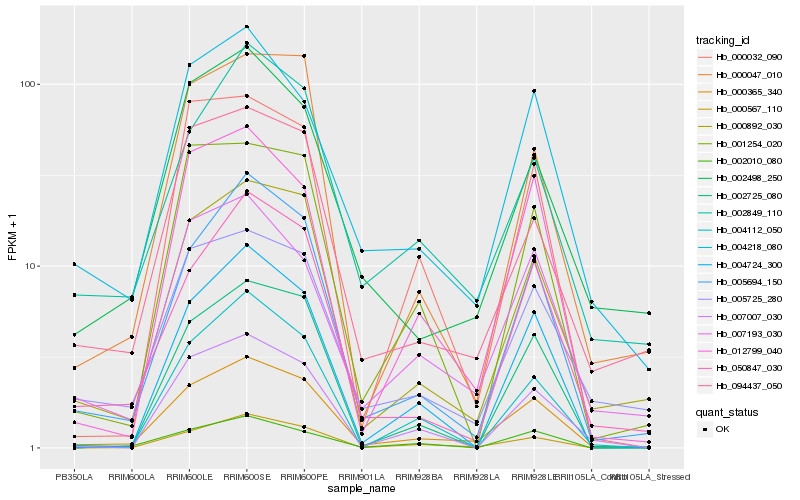
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 2 |
Hb_005694_150 |
0.1050831492 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 3 |
Hb_000892_030 |
0.1072483244 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 4 |
Hb_000567_110 |
0.1199615997 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 5 |
Hb_050847_030 |
0.122256 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 6 |
Hb_012799_040 |
0.1249173932 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 7 |
Hb_004218_080 |
0.1264971529 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 8 |
Hb_007193_030 |
0.1280516923 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005725_280 |
0.1294313296 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 10 |
Hb_002725_080 |
0.1314973095 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_004724_300 |
0.134543695 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 12 |
Hb_007007_030 |
0.1350658134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_094437_050 |
0.1355812171 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002849_110 |
0.1362624808 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 15 |
Hb_000047_010 |
0.138018688 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 16 |
Hb_002498_250 |
0.1434886172 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_004112_050 |
0.1435242904 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_001254_020 |
0.1440476872 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 19 |
Hb_000032_090 |
0.1443589703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002010_080 |
0.1465322275 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |