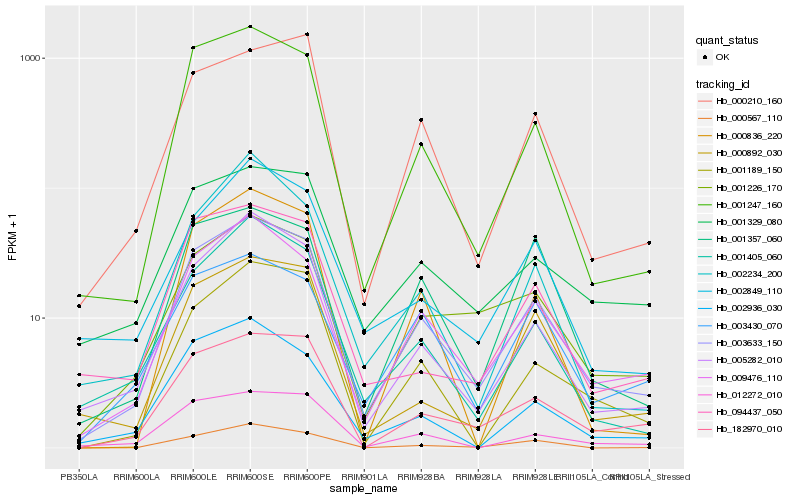
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003633_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 2 |
Hb_009476_110 |
0.0790304613 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 3 |
Hb_001247_160 |
0.0882401657 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 4 |
Hb_005282_010 |
0.1034141731 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 12 [Populus euphratica] |
| 5 |
Hb_182970_010 |
0.1086614064 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 6 |
Hb_000567_110 |
0.1112691774 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 7 |
Hb_001405_060 |
0.1191448384 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |
| 8 |
Hb_001226_170 |
0.1232994224 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 9 |
Hb_001189_150 |
0.1255432773 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 10 |
Hb_012272_010 |
0.1277711553 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_001329_080 |
0.1290272662 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 12 |
Hb_002936_030 |
0.1298129993 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000210_160 |
0.1323069479 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 14 |
Hb_000892_030 |
0.1334632198 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 15 |
Hb_002849_110 |
0.1343623606 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 16 |
Hb_094437_050 |
0.1390950761 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003430_070 |
0.1392572494 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 18 |
Hb_000836_220 |
0.1400943773 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_001357_060 |
0.1410695132 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 20 |
Hb_002234_200 |
0.1425197813 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |